
This web page was produced as an assignment for an undergraduate course at Davidson College.
The Origin of Domestic Dogs

In this study Thalmann et al. researched the origin of domesticated dogs. The goal was to find the location and timeline of modern dog domesticity. This information is of use when trying to understand how a wild carnivore evolved into a human-dependent pet that can now be found practically everywhere humans inhabit. Through the use of molecular dating Thalmann et al. found that the domestication and dispersal of modern domestic dogs has likely been heavily influenced by human growth and population patterns over the past several thousand years.
Prior to this paper there lacked conclusive evidence as to whether the domestic dog originated in Europe or East Asia. Doglike fossils recovered in Europe date back to 30,000 years ago and are the oldest known specimens of the species. Phenotypic variation can be deceiving when examining domestic dogs and other members of the Canus genus, as one can easily mistake physical for genetic variation. For example, comparing the Chihuahua to an American Grey Wolf solely by phenotype could prove tricky. The two dogs are of the same genus, but some might be fair in saying the Chihuahua shares a greater resemblance to those of genus Rattus. By sequencing the mitochondrial genomes of ancient and modern canids, Thalman et al. are able to provide statistically supported data on the genealogy of domestic dogs.
Thalmann et al. compared the genomes of ancient and modern canids and used the information to construct a phylogenetic tree. Mitochondrial DNA was taken from modern wolves (49), modern dogs (77), coyote (4) and 18 prehistoric canids. The modern dog samples included divergent dog breeds and Chinese indigenous dogs. The ancient canid samples were categorized as domesticated as they had phenotypic traits indicative of domesticity. Statistical analysis (maximum likelihood, coalescence, Bayesian) of the results was done to construct the phylogenetic tree that represents the most likely relation of all of specimens.
The phylogenetic tree divided all modern dogs into four clades; A,B,C,D. Clades A, C, and D share significant sequence similarity with ancient European canid specimens. The other modern dog clade, B, shows similarity with modern European wolves. This data suggests that all four clades of domestic dog originated in Europe. Furthering the support for the European model of origin, when comparisons of modern wolves from the Middle East and East Asia were done against the modern clades, no significant similarities were found. Dog-wolf divergence was also determined to most likely have taken place 15,000 years ago. This would place the domestication of dogs somewhere within the Last Glacial Maximum. Timing of domestication suggests that domestication may have been taking place as the earliest human hunters were at work. The hypothesis that Thalmann et al. offers is that proto-dogs may have fed on the carcasses left behind by humans, provided aid in the hunt, or offered defense.
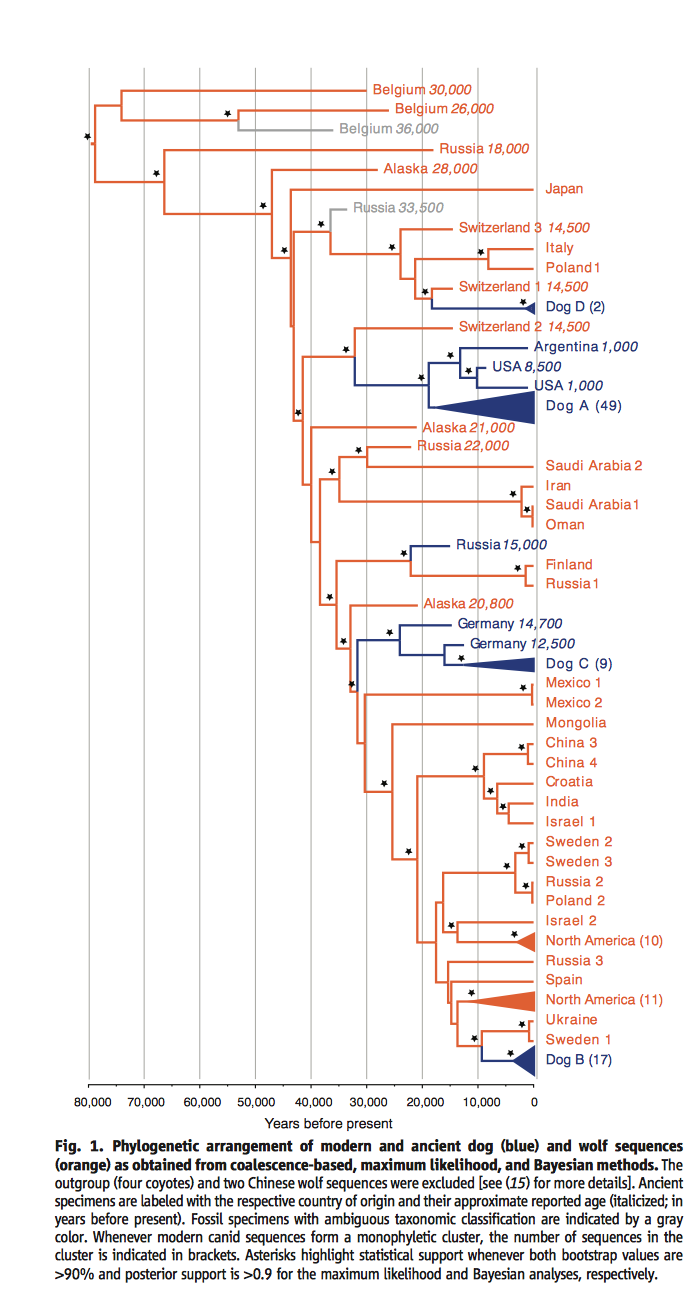
Figure 1.
This phylogenetic tree above was constructed using data provided by mitochondrial genome sequencing. The tree includes both modern dogs (blue) and ancient dogs (blue), as well as present and ancient wolves (orange). The asterisks at diverging points on the tree show that there is statistical support that bootstrap values from the maximum likelihood approach are greater than 90% and posterior support is greater than .9 from Bayesian analyses.
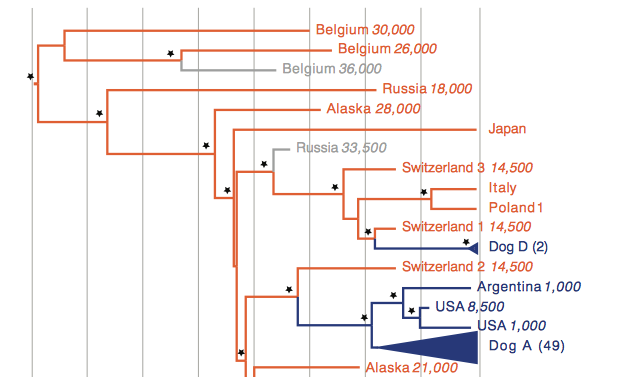
In this portion of Fig. 1. one can see that Belgium 30,000 is the oldest canid of the tree. Category name is assigned by the location where a sample was found and how many years ago from present the specimen it was dated to. Belgium 36,000 and Russia 33,500 are both shown in gray as their classification as dog or wolf is unclear. The modern dog clade Dog D is the least diverse of the four modern clades. As shown, Dog D is distinguished as its own clade based on two modern dog samples. These dog samples are from two different Scandanavian breeds. These two breeds are sister to a wolf canid in Switzerland. Dog Clade A is the most diverse of the modern dog clades as it contains the majority of modern dog sequences. What is interesting about Dog A and the New World dogs (Argentina 1000, USA 8500, USA 1000) is that they share a MRCA from 18,000 years ago. This suggests that dogs in the pre-Colombian era share ancestry with modern dogs. Looking back further, the New World dogs and modern dogs have a MRCA with Switzerland 2 14500. This might mean dogs reached the New World by following human migration from Eurasia, specifically coming from Switzerland.
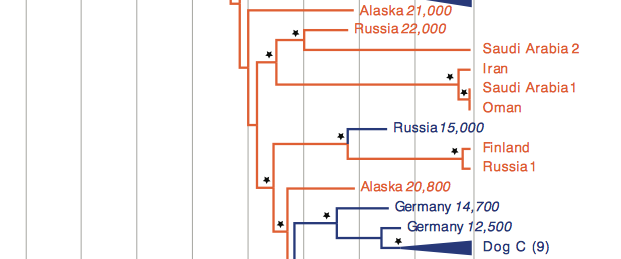
In this second portion of Fig. 1. shown above, one can see several different wolf samples, Dog C, as well as three different ancient dogs. Russia 15000, Germany 14700, and Germany 12500 are all ancient domesticated dog specimens that did not survive.This is of interest as it indicates domesticity may have taken place in multple circumstances. Though these two specimens are exmaples of where domesticity failed. Dog C contains 12% the sequences collected from modern dogs. This modern clade is most closely related to ancient dog species recovered in Germany.
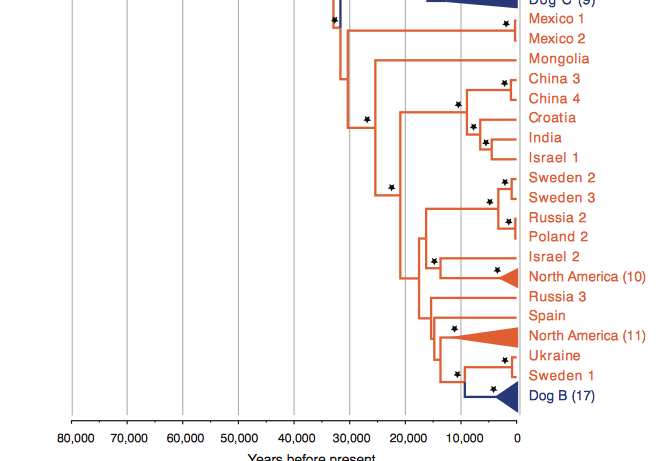
In the bottom third of Fig. 1. modern wolf specimens and Dog B clade can be seen. Modern dogs from clade Dog B can trace their origin back to wolves in Europe. All of the modern wolves from the Middle East/East Asia sequenced (Israel 1, China 3 and 4, Mongolia) do not have a close relation with Dog B. The fact that Dog B shares a MRCA with wolves of European origin supports the European hypothesis for dog domesticity.
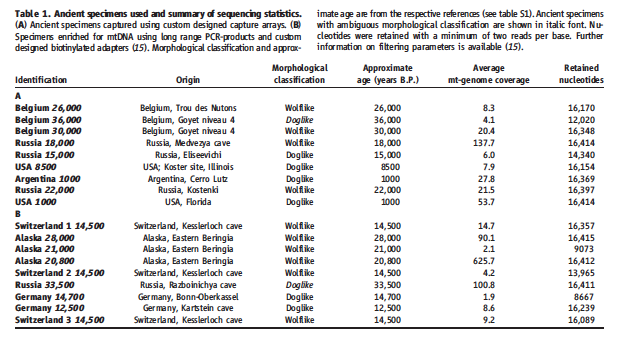
Table 1.
Table 1. provides information on the ancient dog and wolf samples whose mitochondrial DNA was sampled and used in genome comparisons. Identification as A or B indicated the method by which mtDNA was captured. Group A specimens were captured by custom designed capture arrays. MtDNA from B specimens were captures using long range PCR-products. Morphological classification as Wolflike or Doglike is also included. Approximate age, average genome coverage, and the number of retained nucleotides is also given for each specimen.
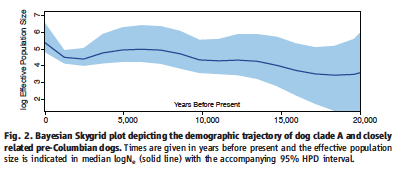
Figure 2.
Fig. 2. is a Bayesian Skygrid that gives the predicted population size of the modern dog clade Dog A and pre-Columbia dogs over the past 20,000 years. Population growth is seen in this group from 20,000 years before present until 5,000 years before present. At this point the population size stagnated and then dipped. Population growth began again around 2500 years ago and has continued to be on the rise. This trend mimics human demographic changes over the same period.
I applaud this paper for its clear charts and figures. They did a good job of showing the most relevant information in their figures. They were also thorough in explaining their data and its implications in the literature. What the paper did lack was a good explanation of their three different statistical methods. Given that this was the basis for constructing their entire tree and argument, I would have liked to be given more information. In Table 1. they also mentioned the use of two different methods of DNA capture, but they fail to explain this in the literature. I also want an explanation as to how they chose what modern dogs to samples and if they tried to extract mtDNA from all recovered ancient canid fossils avaible. I feel like this paper was concise and clear in their explanations of their data. However, I want more information on their methods which I think is an important component lacking in this paper.
References:
Thalman et al. Complete Mitochondrial Genomes of Ancient Canids Suggest a European Origin of Domestic Dogs. Science 342: 871-874 (2013).
Genomics Page
Biology Home Page
© Copyright 2014 Department of Biology, Davidson College, Davidson, NC 28035