This web page was produced as an assignment for an undergraduate course at Davidson College.
Nick's Home Page Genomics Page Biology Home Page
Assignment 1
New genomics tool could help predict tumor aggressiveness, treatment outcomes
Intra-tumor Genetic Heterogeneity and Mortality in Head and Neck Cancer: Analysis of Data from The Cancer Genome Atlas
1) What was the research project?
This
focused on developing a method to quantify the heterogeneity of
individual cases of head and neck cancer and correlating increased
intra-tumor variation with higher rates of mortality using a newly
developed mutant-allele tumor heterogeneity (MATH) statistic.
2) Were they testing a hypothesis or doing discovery science?
This research was based on a standing, but untested, hypothesis that tumors containing high genetic diversity were more resistant to treatment and more aggressive to hosts. This could possibly be due to different intra-tumor populations being resistant to different types of treatment, making single-method treatment much less effective.
3) What genomic technology was used in the project?
Publically available clinical data and whole exome sequence data (sequences from all protein-producing genes) from over 300 patients were used in this study and obtained from The Cancer Genome Atlas project. Researchers were able to use cancer genomes paired with healthy genomes from the same patient to compare the frequency of mutant alleles in populations and subpopulations of tumor cells. In doing so, they were able to show a strong correlation between mortality and increased heterogeneity, even when other clinical (HIV, smokers, etc.) or molecular (TP53 mutation) factors were accounted for.
4) What was the take home message?
Increased genetic diversity, as represented by the MATH statistic, in tumors correlate with higher patient mortality in head and neck cancers. In a clinical setting, increased heterogeneity could make targeted cancer treatment more difficult, since it would be equivalent to treating two or more cancers simultaneously.
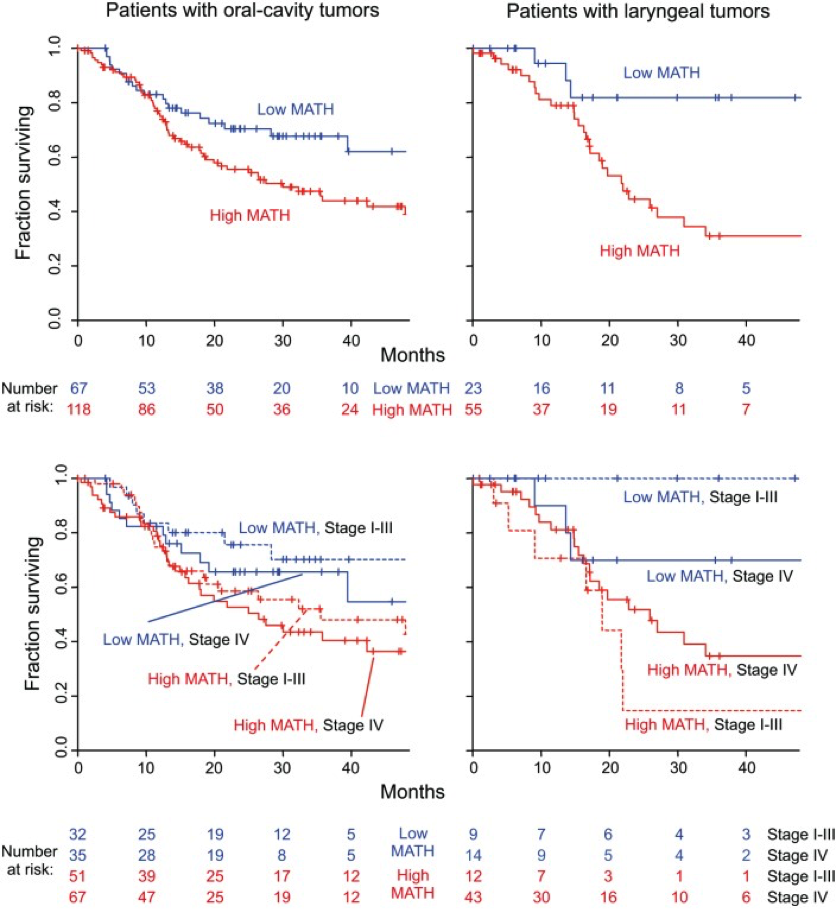
Figure 7. MATH value and mortality in patients with tumors in the oral cavity or the larynx. Left, patients with oral-cavity tumors; right, patients with laryngeal tumors. Top, Kaplan-Meier curves for all patients. Blue, low MATH; red, high MATH (MATH > 32). Cox proportional hazards analysis: oral cavity, high/low MATH HR, 1.69 (95% CI, 1.04 to 2.73; p = 0.033); larynx, high/low MATH HR, 3.55 (95% CI, 1.25 to 10.1; p = 0.018). Bottom, joint relation of MATH value and disease stage to survival; dashed, lines Stages I–III; solid lines, Stage IV. Cox proportional hazards analysis: oral cavity, high/low MATH HR, 1.67 (95% CI, 1.03 to 2.70; p = 0.037, Wald test); Stage IV/Stage I–III HR, 1.34 (95% CI, 0.87 to 2.08; p = 0.19); larynx, high/low MATH HR, 3.50 (95% CI, 1.18 to 10.4; p = 0.024); Stage IV/Stage I–III HR, 1.04 (95% CI, 0.44 to 2.49; p = 0.92). (replicated from Mroz et al., 2015)
I thought that this figure (Figure 7 from the
paper) is effective at visually representing the importance of the
calculated MATH scores for patients. The top figures represent the
rate of survival of patients with high and low MATH scores, regardless
of the stage of progression of the cancer. The bottom figures divide
the cancer groups into stages of progression and show that low MATH
scores are a better indication of survival after treatment than the
stage of the cancer at treatment. It is important to note that the
right graphs have a lower sample size, skewing the data to have
seemingly more drastic outcomes.
More broadly, this highlights the genomic mosaicism
that can be present in individuals, where the genome of a specific
cell may not exactly match the genome of another cell in the same
organism.
5) What is your evaluation of the project?
I thought that this was a very good example of a hypothesis-driven
genomics study. The researchers were able to use publicly available
data, and, moreover, they were able to use whole exome sequencing
data, which is, I think, a more likely option for further clinical
prognosis due to smaller size and, therefore, less cost. While the
statistic that they came up was verified on other cancer types, to
could prove to be a useful tool for doctors to evaluate how best to
treat a cancer and how quickly they need to act on a cancer. The
summary sheds good light on what the goal of the research was and
highlights some of the major findings, though a few of the figures
from the original paper would have been easy to incorporate and
interpret, even without a scientific background, Figure 7 above. I
would like to see research done to apply this statistic to other
cancer types and especially to younger groups of patients (mean age in
the study was approx. 61, likely due to the nature of the cancer and
how it is related to activities such as smoking).
Citations
Mroz,
E. A., Tward, A. M., Hammon, R. J., Ren, Y., & Rocco, J. W. 2015.
Intra-tumor genetic heterogeneity and mortality in head and neck
cancer: analysis of data from The Cancer Genome Atlas
[Internet]. 2015. PLoS Medicine. [cited 27 Jan 2016]; 12(2),
e1001786. Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC4323109/.
Ohio State University Wexner Medical Center. New genomics tool could help predict tumor aggressiveness, treatment outcomes [Internet]. 2015. ScienceDaily; [cited 2016 Jan 27]. Available from: http://www.sciencedaily.com/releases/2015/04/150416192745.htm.
Nick's Home Page Genomics Page Biology Home Page
Email Question or Comments to nielder@davidson.edu
© Copyright 2016 Department of Biology, Davidson College, Davidson, NC 28035