This web page was produced as an assignment for an undergraduate course at Davidson College

Figure
1:
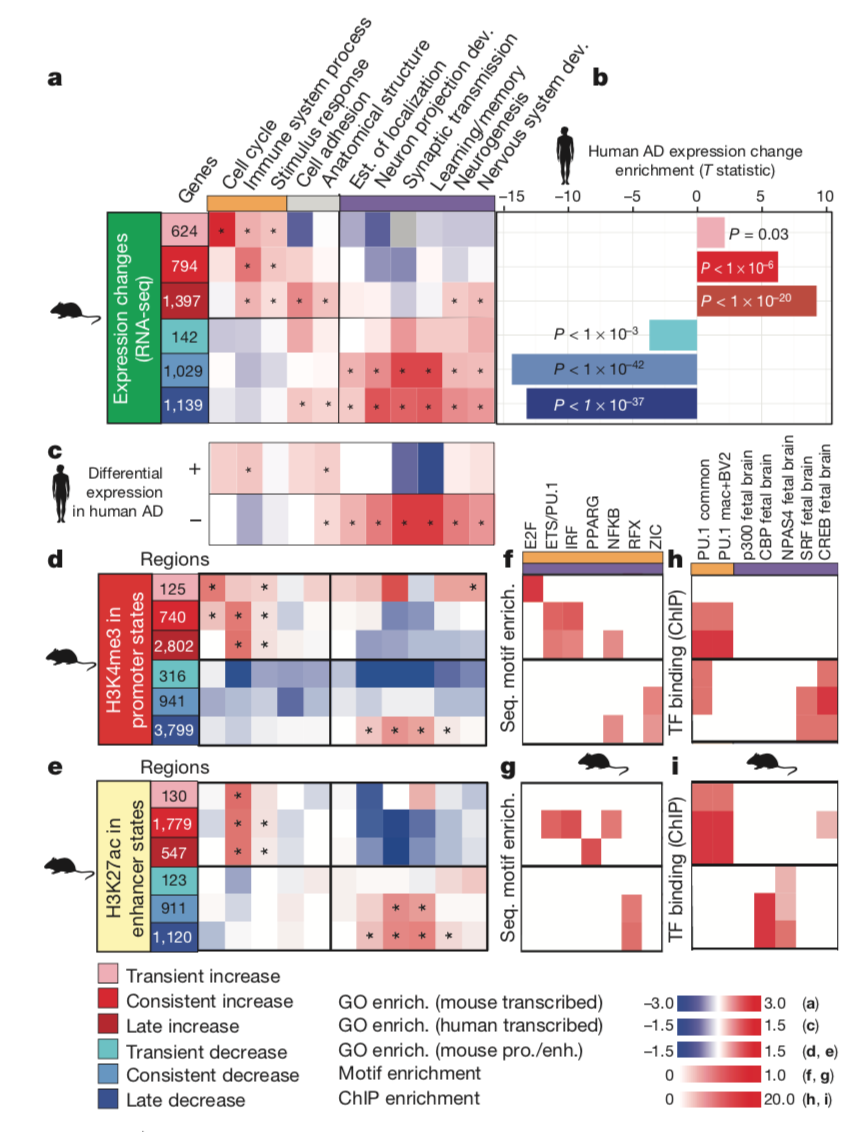
This figure shows gene expression changes in both mice humans with
Alzheimer's disease. Panel a depicts changes in gene expression for six
different temporal classes in eleven of the most significant Gene
Ontology categories. Panel b corresponds to Panel a in that it shows the
T-statistic of gene expression change for each temporal class, but this
time in humans with Alzhemier's disease instead of in mice. Panel c is
supposed to depict human gene expression change per temporal class in
patients with Alzheimer's disease compared to a control, but it is
unclear where the control data versus patient data are in the panel.
Panels d and e show promoter states and enhancer states in mice, and the
enrichment of each Gene Ontology category was calculated with respect to
those states. Panels f and g show the enrichment of the regulatory
motifs in promoters and enhancers, which are the top and bottom of each
panel respectively; Panel f is a control, and Panel g is conducted with
mice modeling human Alzheimer's disease. Panels h and i show where
promoters and enhancers within the mouse models overlap with neuronal
and immune transcriptional factors, showing that gene expression changes
in humans with Alzhemier's disease and the mouse models are related to
immune and neuronal funcions.