pBR322 PCR
BACKGROUND
The polymerase chain reaction (PCR) was first developed in 1985. Quickly, it
became one of the most used and useful
tools available to molecular geneticists. As we have and will discuss in class,
PCR has been used for a host of purposes. The importance of this technique to
biomedical research was illustrated when Kary
Mullis, the inventor of PCR, received the Nobel Prize in Chemistry in 1993
Basically, the polymerase chain reaction is an in vitro enzymatic technique which results in the exponential amplification of a targeted DNA sequence. The process consists of three main steps: denaturation, annealing, and extension.
Denaturation — double stranded DNA must be denatured so that each strand can serve as a template for the production of a complementary strand. Denaturation typically is achieved by heating DNA to 95oC.
Annealing — entails addition of two specific oligonucleotide primers and their subsequent hybridization to target sequences within the denatured DNA. Annealing temperature depends on sequence and length of primers.
Extension — DNA synthesis. DNA polymerase adds new nucleotides to free 3’ ends of annealed primers, Temperature must be compatible with activity of enzyme.
For a primer on PCR, click here.
Oligonucleotide Primers
Generally, primers are 18-24 bases in length.
Approximately 50% GC content.
Both primers used should have similar Tms.
Annealing temperature generally 5oC below Tm.
Primer should contain little internal homology.
Primers should contain a 3’ G or C
Generation of degenerate primers.
Degeneracy of genetic code
Codon usage tables for different species
Thermostable DNA polymerases
Taq — originally isolated from
Thermus aquaticus.
Possesses 5’-3’ DNA synthesis activity.
Lacks 3’-5’ exonuclease (proofreading) activity.
VENT — isolated from Thermococcus litoralis.
Possesses proofreading activity.
Pfu — originally isolated
from Pyrococcus furiosus.
Possesses proofreading activity.
Variations on a theme
Basic PCR — amplification of DNA using
two primers complementary to ends of region to be amplified
RT-PCR — combined use of reverse transcription and PCR to amplify mRNA
PCR Cloning — primers designed to incorporate restriction enzyme sequences
at their 5’ ends to facilitate cloning
PCR-based Site-directed
mutagenesis — primers designed to incorporate a specific nucleotide
mutation, thereby producing a mutated PCR product
GOALS
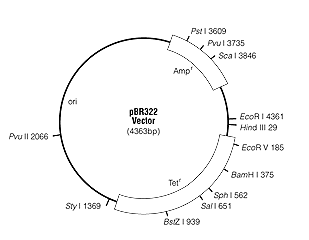
Amplify tetracycline resistance gene from pBR322
plasmid
Confirm identity of PCR product by gel electrophoresis and Southern blot
Ligate PCR product into another plasmid
Transform tetracycline sensitive E. coli with the recombinant plasmid
Confirm phenotype of transformed bacteria
MATERIALS
Template DNA: pBR322

Primer 1: 5’-CGCAGTCAGGCACCGTGTATG-3’
Primer 2: 5’-CCATTCAGGTCGAGGTGGCCC-3’
To a 0.5mL microfuge tube, add the following reagents:
| Reagent (Stock Concentration) | Volume | Final Concentration |
| Deionized water | 6.5 ul | |
| PCR Mix (2x stock) | 12.5 ul | 1x |
| Primer 1 (10 uM) | 2.5 ul | 1.0 uM |
| Primer 2 (10 uM) | 2.5 ul | 1.0 uM |
| pBR322 DNA (0.25 ug/uL) | 1 ul | 5 ng/uL |
NOTE: The PCR mix contains Taq DNA polymerase, dATP, dTTP, dCTP, dGTP, and a buffer.
Briefly centrifuge the tube to collect all material
at the bottom.
Cycle as follows:
94oC 2 minutes — 1
cycle
94oC 1 minute
50oC 1 minute
72oC 1 minute
30 cycles
Store amplified DNA at —20oC.