|
Microbes Fight Back Against
Antibiotics
|
|
|
Microbes, have developed
complex tools to combat antibiotics. Many bacteria have evolved
efflux pumps, which transport antibiotics out of the organism before
the drug can be effective. The antibiotic tetracycline, for example,
must enter the cell and attach to the 30s ribosomal subunit in order
to inhibit bacteria. Tetracycline resistant bacteria, however, remove
the drug from the organisms before it can attach to the ribosome,
thereby withstanding up to 100 times the therapeutic dosage of susceptible
bacteria. Microbes have also evolved enzymes, which degrade the
drug before it reaches the target within the microbe. This mechanism
of resistance is extremely effective and the most widespread of
any of the modes of resistance. Enzymes alter, and so inactivate,
many antibiotics, including streptomycin, kanamycin, cephalosporin,
and penicillin. Another, less common form of resistance, is the
alteration of the antibiotic target within the bacteria. Instead
of attacking the antibiotic, some bacteria alter the chemical structure
of the target yielding several classes of antibiotics useless. Using
this mechanism, resistance to rifampin, quinolones, and tetracylclines
have evolved in many classes of bacteria. Thus, bacteria have evolved
numerous mechanisms to combat common antibiotics.
|
|
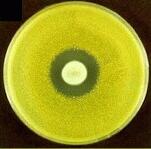
| The fungus Penicillium notatum
was allowed to grow on this plate the yellow bacterium, Micrococcus
luteus was overlayed. The absence of yellow in the center of the
plate shows the antibiotic effects of penicillin on this bacteria.
Image from The
University Edinburgh. |
| Transmitance of Resistance
Genes |
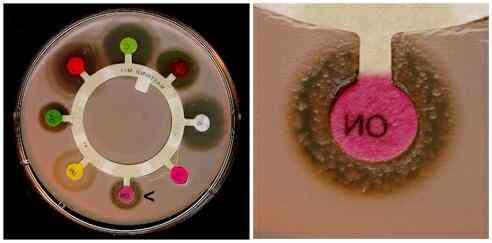
| This plate
analyzes the resistance of various antibiotics against the Bacillus
bacteria. Bacillus bacteria that are affected by the antibiotic
will not grow around the circle of antibiotic. However, the
novobiocin antibiotic, identified with the arrow in the whole-plate
image, is not effective against Bacillus because the bacteria
has developed resistance to the antibiotic. Image from The
University of Edinburgh. |
|
The dilemma of antibiotic
resistance is heightened by the numerous methods bacteria have evolved
to spread resistance genes. On occasion, bacteria develop a point
mutation that resists antibiotic treatment. The resistant gene can
then be passed down to progeny to create more resistant bacteria.
This process is called vertical genetic exchange. More frequently,
microbes use horizontal genetic exchange to transmit resistant genes
to bacteria other than the progeny.
|
|
Horizontal genetic exchange
takes place by conjugation, transposition, transduction, and transformation.
Conjugation occurs between two bacteria, one containing the resistance
gene and one susceptible to the therapy. The resistance genes, encoded
on small circular pieces of DNA, are copied and transferred to the
susceptible bacteria. Microbes also spread resistance genes by transposition.
In this process transposons, small segments of DNA, jump from bacteria
to bacteria, each time leaving a copy of the resistance gene. Transduction
is another frequent mechanism of resistant gene transfer. Transduction
occurs when a bacteriophage, a virus specific for bacteria, carries
a resistant gene. Upon infection of the bacteria the resistant gene
can be incorporated in the bacterial genome, thus causing resistance.
Another, method of resistant gene transfer is transformation. Transformation
takes place when free DNA, encoding antibiotic resistance, is incorporated
into the genome of bacteria. Thus, through different, and integrative
transfer processes bacteria have evolved to resist many of the current
antibiotics.
|
|

