RT-PCR
(Reverse Transcriptase Polymerase Chain Reaction)
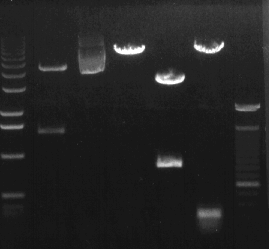
permission requested from http://mcbio.roswellpark.org
This page was created as part of an undergraduate assignment at Davidson College.
|
RT-PCR (Reverse Transcriptase Polymerase Chain Reaction) |
permission requested from http://mcbio.roswellpark.org |
WHAT IS IT?
RT-PCR stands for Reverse Transcription-Polymerase Chain Reaction. It is a technique used in genetic studies that allows the detection and quantification of mRNA. It is a very sensitive method that shows whether or not a specific gene is being expressed in a given sample. RT-PCR is a very important test in the field of Genetically-Modified Organisms (GMO's) because it gives researchers a mechanism to test whether any specific gene is turned on (active) or turned off (inactive). This allows researchers to identify the benefits of genetically-modified organisms with respect to their "natural" counterparts and search for any significant differences in which genes are expressed in the two types of organisms.
HOW DOES IT WORK?
RT-PCR is used to locate and quantify known sequences of mRNA in a sample. The first step in RT-PCR uses reverse transcriptase and a primer to anneal and extend a desired mRNA sequence. If the mRNA is present, the reverse transcriptase and primer will anneal to the mRNA sequence and transcribe a complimentary strand of DNA. This strand is then replicated with primers and Taq Polymerase, and the standard PCR protocol is followed. This protocol copies the single stranded DNA millions of times in a small amount of time to produce a significant amount of DNA. The PCR products (the DNA strands) are then separated with agarose gel electrophoresis. If a band shows up for the desired molecular weight, then the mRNA was in fact present in the sample, and the associated gene was being expressed.
WHAT DOES IT TELL US?
The results from RT-PCR are used in two main ways. First, RT-PCR shows us whether or not a specific gene is being expressed in a sample. If a gene is expressed, its mRNA product will be produced, and an associated band will appear in the final agarose gel with the correct molecular weight for the gene. This is used in GMO research to identify whether or not a transplanted gene is actually working within the new organism. Also, RT-PCR can quantify exactly how active the gene is in the sample. To do this, RT-PCR is performed with the unknown mRNA alongside standardized samples with known mRNA amounts. This approach is used to identify how much mRNA is being produced by the gene. This technique is especially useful in comparing a GMO to its natural counterpart when the modification causes an upregulation or downregulation of a specific gene (for example, vitamin content in "golden rice"). These two ways of using RT-PCR are important tools in the production and design of GMO's. In addition, RT-PCR is a crucial method after production of the GMO when proving to the public that GMO's are safe and marketable alternatives to their natural counterparts.
LINKS TO OTHER RT-PCR WEBPAGES
http://www.alkami.com/methods/mthdrt.htm#literature
http://www.alkami.com/qguide/Qguide-3.pdf
http://bio.davidson.edu/courses/Immunology/Flash/RT_PCR.html
http://www.promega.com/guides/pcr_guide/070_12/promega.html
http://www.ambion.com/basics/rtpcr
http://www.liv.ac.uk/~giles/methods/rlpcr.htm
BIBLIOGRAPHY
Campbell, A. Malcolm. "RT-PCR Methodology." Davidson College, Immunology Methods. 2000. 9/9/02. <http://bio.davidson.edu/courses/Immunology/Flash/RT_PCR.html>
"Protocols and Reference - Introduction to RT-PCR." Promega Corporation. 2000. 9/9/02. <http://www.promega.com/guides/pcr_guide/070_12/promega.html>
"Reverse Transcriptase PCR." Alkami Biosystems. 7/10/02. 9/9/02. <http://www.alkami.com/methods/mthdrt.htm#literature>
"RT-PCR - The Basics." Ambion, Inc. 2002. 9/9/02. <http://www.ambion.com/basics/rtpcr>
 I'D LIKE AN A FOR THIS WEBPAGE
I'D LIKE AN A FOR THIS WEBPAGE

© Copyright 2002 Department of Biology,
Davidson College, Davidson, NC 28035
Send comments, questions, and suggestions to: pelowry@davidson.edu