Our Research


Emily Oldham and Dan Pierce ('03)
How do you know if what you are measuring accurately represents your sample?
Experimental Design Questions
Define probe v. targetLabel RNA v. cDNA (impact on spotted material)Cy series v. Alexa series dyesSpots made of oligos v. cDNAAffymetric chips v. printed glass chipsReproducibility within and between labs
Controls
Dye reversalPooled v. control reference mRNAControl spots from another species
Our Research


Emily Oldham and Dan Pierce ('03)
How do you know if what you are measuring accurately represents your sample?
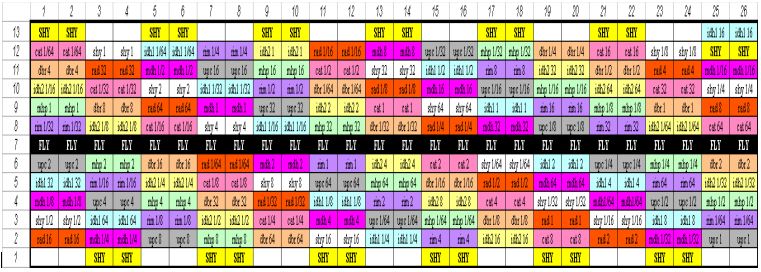
Layout of our DNA Microarray
Experimental Design
Need Stronger Signal
3DNA Dendrimer Advantage = 300+ dyes per binding
DNA Concentrations Printed
Establish Scanning Parameters
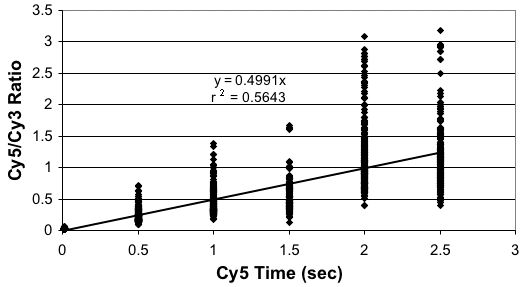
Concentrations Alters Ratios
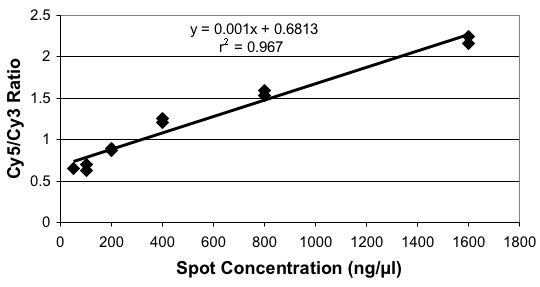
Sequence-Specific Variations
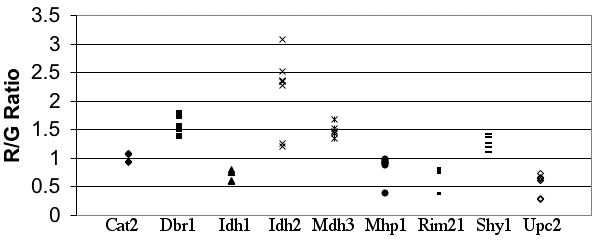
Altered 3DNA Sticky Ends
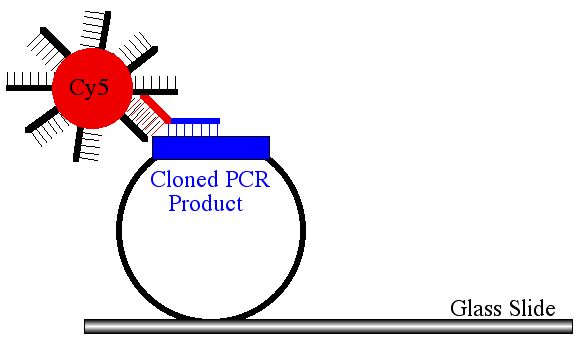
Wanted to Improve Quality of Signal: Clone PCR Products
Original PCR Products |
Cloned PCR Products |
 |
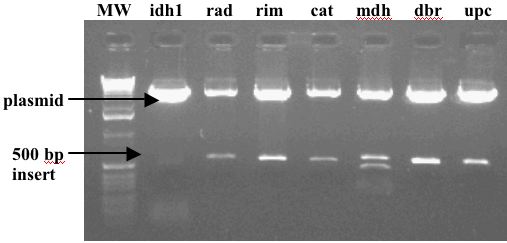 |
Clean Up Printed DNA: Clone PCR Products
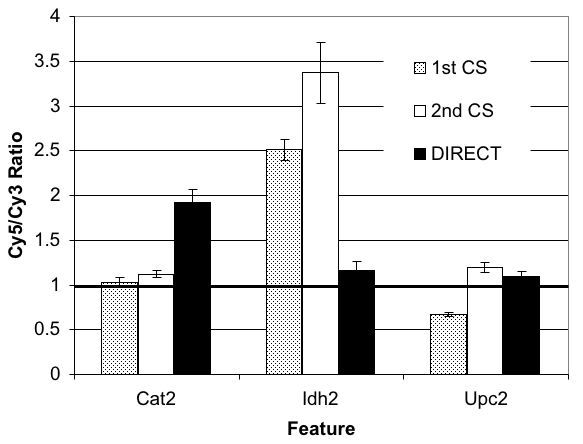
Optimized Experimental Conditions: Signal Up and Noise Down
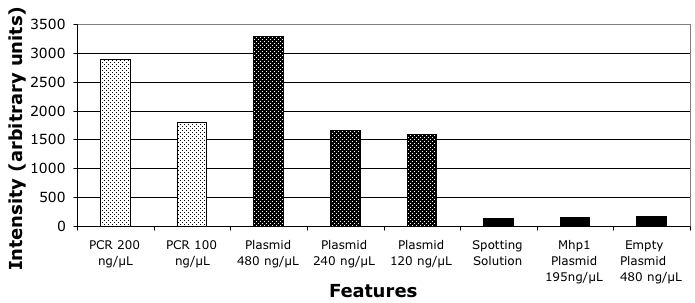
Plasmids Improve Signal
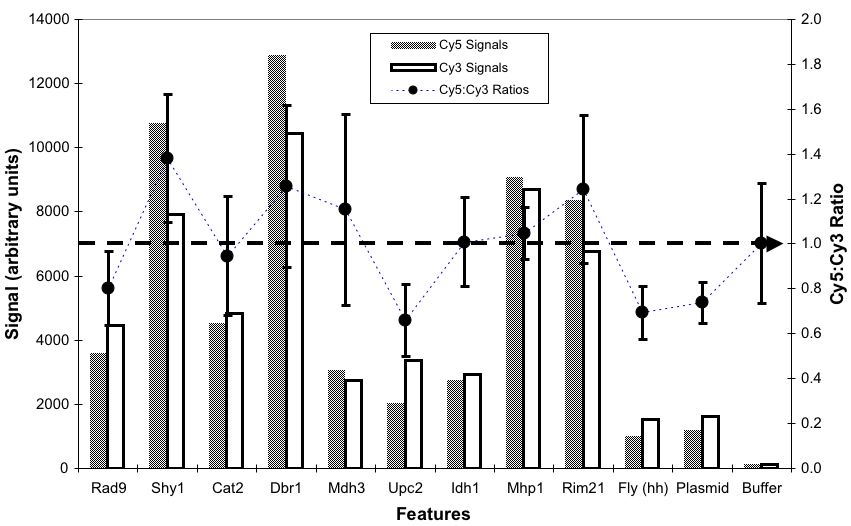
What about Other Ratios? 10:1, 3:1,
1:1, 1:3, 1:10
(expected log2 = 3.3, 1.6,
0, -1.6,
-3.3)

Good Trends