This
page was produced as an assignment for an undergraduate course at Davidson
College.

Telomerase
A SOLUTION TO THE "END-REPLICATION" PROBLEM (click here for a review of DNA replication)
- Because DNA polymerases operate only in the 5' to 3' direction, synthesis of the lagging strand occurs through a "backstitching" mechanism that produces short fragments of DNA. This technique causes a problem at the end of a linear chromosome: there isn't enough room for the RNA primer needed to start the last fragment at the chromosome's 3' end. Without its complement, the hanging piece of unpaired DNA from the parent strand might be recognized by the cell as a broken piece of DNA and then damaged or even chopped off in the cell's attempt to mend it.
- To prevent the "correction" of the unpaired but important piece of DNA, eukaryotes use certain G-rich nucleotide sequences as protective "caps." Called telomeres, these caps incorporate many tandem repeats of the nucleotide sequences (human telomeres extend for about 10,000 nucleotides), ultimately allowing efficient replication of the chromosome ends (Alberts et al, 2002).
- Telomeres are created by telomerase, a riboprotein complex that uses its own piece of RNA primer to elongate the chromosome ends until the DNA polymerase can move in and complete the lagging strand.
WHAT DOES TELOMERASE DO?
- When a chromosome is replicated, the 3' hanging parental fragment containing the G-rich sequence GGGTTA (in humans) is recognized by the telomerase complex. Using its own RNA as a complemantary primer, the telomerase mimics reverase transcriptases and elongates the hanging DNA fragment in the 5' to 3' direction, moving down the growing strand until it is a telomere several thousand bases longer. Replication of the lagging strand can now be completed using the telomeric extensions as a template for synthesis using DNA polymerase (Alberts et al, 2002).
- With this mechanism, of course, the 3' end of each telomere is still slightly longer than the complementary 5' end. Other telomere-associated proteins move in to loop the protruding end back on itself, protecting it from degrading enzymes and other mechanisms for repairing broken DNA.
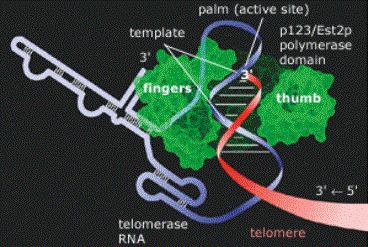
Figure 1: Telomerase Structure at Work. The
telomerase enzyme complex consists of an RNA component and a protein component.
The protein component's catalytic subunit acts as a reverse transcriptase
(green), using telomerase RNA (blue/white) as a template for the addition of
telomeric repeat sequences to the telomere DNA strand (red).
Source: Thomas Cech
TELOMERASE STRUCTURE AND HOMOLOGS
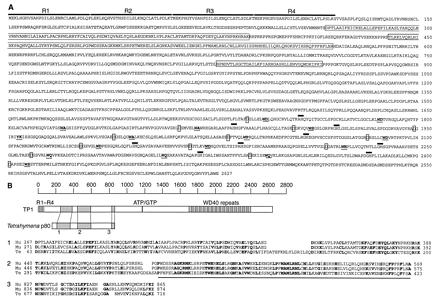
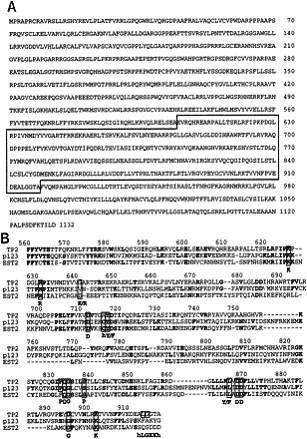
- TERT, the gene for the telomerase catalytic subunit (the reverse trascriptase mimic) is approximately 4 kb in length and exists somewhere in the chromosome as a single-copy gene (Kilian et al, 1997). The catalytic subunit, named TP2, has a molecular weight of approximately 130 kDa. Shaped almost like a hand as seen in Figure 1, its "fingers" and "thumb" pull the telomerase RNA and telomere DNA strands into the active site (the "palm"), where the telomere is elongated. The primary structure (amino acid sequence) of TP2 is shown in Figure 2a (Harrington et al, December 1997).
- The other major telomerase protein is the RNA-binding subunit, named TP1; its amino acid sequence is shown in Figure 3a. The amino-terminus of TP1 is sufficient to bind telomerase RNA in vivo (Harrington et al, January 1997).
- Both TP1 and TP2 have similarities with telomerase-binding proteins of other species. The amino-terminus of TP1 is both an RNA binding site and the sequence segment most homologous to Tetrahymena p80, a ciliate RNA-binding protein, as seen in Figure 3b, and Figure 2b demonstrates the amino acid sequence similarity between the Est2 (yeast), p123 (ciliate), and TP2 telomerase catalytic subunits. These results suggest that the components of the telomerase complex are evolutionarily conserved as homologous structures.
(click picture to see larger image)
Figure 2: Amino acid sequence of human TP2.
(a) Predicted amino acid sequence of human TP2. Homology to the
reverse transcriptase domain is boxed with a solid line. (b) Domain of
highest homology between TP2 (human telomerase catalytic subunit), p123
(ciliate Euplotes aediculatus ~123 kDa telomerase catalytic subunit),
and Est2 (yeast Saccharomyces cerevisiae homologous
telomerase-associated subunit). Residues conserved in at least two of the
aligned proteins are shown in boldface type. Residues conserved among
reverse transcriptase are boxed, with consensus residues shown below.
Source: Harrington
et al, December 1997
(click picture to see larger image)
Figure 3: Amino Acid Sequence of Human TP1. (a)
Four amino-terminal repeats are indicated by solid lines over the sequence (R1
to R4). Boxed regions show homology to Tetrahymena p80. The
complete human and murine TP1 sequences have been deposited with GenBank
(accession numbers U86136 and U86137, respectively). (b) (Top)
The TP1 amino acid sequence is depicted schematically, with boxed regions as
follows: amino-terminal repeats 1 to 4 (R1-R4); gray boxes (1 to 3) show
regions of homology between TP1 and Tetrahymena p80. (Bottom)
Human (Hu) and murine (Mu) TP1 amino acid sequences were aligned to Tetrahymena
p80; Tetrahymena (Te) sequences identical to either human or mouse at
each position are shown in boldface. The underlined, 90-amino acid segment in
region 2 contains 46% identity between p80 and human TP1.
Source: Harrington
et al, January 1997
- In addition to the telomerase protein components showing similarities, the telomerase RNA itself shows signs of being at least partially conserved. The TP1 and p80 similarities at the amino-termini suggest that at least the telomerase-binding sites must be similar between the two species' RNA strands. As Figure 4 shows, possible secondary structures of the two species' RNA may be similar enough to be recognized by an evolutionarily conserved RNA-binding motif.
 a
a
 b
b
Figure 4: A Comparison of Human and Yeast/Ciliate
Telomerase RNA. Vertebrate/human telomerase RNA (a) and yeast/ciliate
telomerase RNA (b) differ greatly in sequence and structure, but they share a
5' pseudoknot area close to the template sequence (i.e. the portion used to
generate the telomere).
Source: (a) Rfam
3.0 Telomerase-vert
(b) Rfam 3.0
Telomerase-cil
LOCATING TELOMERASE IN THE GRAND SCHEME OF THINGS
- Telomerase is found in the nucleus, where DNA replication takes place. Not all cells, however, show evidence of telomerase activity. Expression of TERC, the gene for the RNA component of telomerase, is widespread, but the expression of the protein component's catalytic subunit, TERT, is restricted to locations of telomerase activity. Telomerase activity was found to be almost absent in the majority of normal adult tissues, including cardiac and skeletal muscle, adipose tissue, lung, liver, and kidney (Kolquist et al, 1998).
- Because of this curious lack of telomerase activity, a theory arose connecting telomere length to aging and cell senescence. According to this theory, human somatic cells are born with a full number of telomeric repeats, but the telomerase enzyme is "turned off" in some tissues. The cells of those tissues would lose about 50 to 100 nucleotides from each chromosome end each time they underwent replication and division. Eventually, the telomeres would cease to exist and the chromosomes themselves would start losing nucleotides, carrying genetic defects into their next division so that neither daughter cell would be viable and would consequently undergo senescence (Alberts et al, 2002). Thus after a certain number of divisions a cell has "aged" and died.
- This theory has been tested using human fibroblasts grown in tissue culture. Human fibroblasts normally divide around 60 times before dying, and like most other somatic cells, they do not produce telomerase. Their telomeres gradually shorten as they continue dividing. When the telomerase gene is added to the cells' genomes, however, they maintain their telomere length, and some of the cells are able to proliferate indefinitely.
- If telomere length is a cause of human aging, then, might exposure to telomerase lengthen one's life?
- A control on cell division certainly seems important, however, in light of the fact that approximately 85% of all tumors show evidence of telomerase activity (Kilian et al, 1997). If telomerase is provided to maintain telomere length, a cell's life may be prolonged indefinitely, causing genetic instabilities that in turn lead to variant, cancerous cells. Figure 5 compares telomerase gene expression in normal human tissue and concerous tissue, suggesting the correlation.
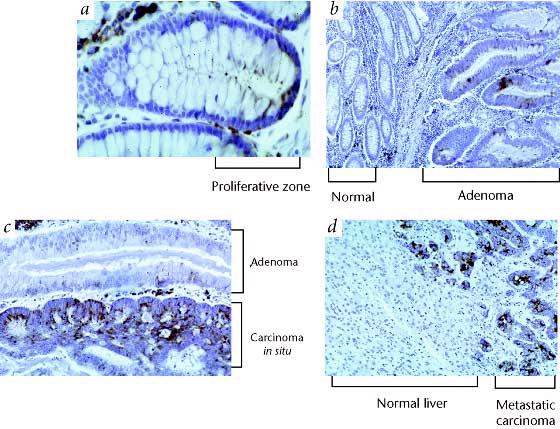 `
`
Figure 5: TERT Expression in Colonic Tumorigenesis.
(a) Normal colon with low-level hybridization to epithelial cells
located primarily in the proliferative zone near the base of crypts. (b)
Increased expression in adenomatous epithelium (right), normal mucosa is on the
left. (c) Increased expression in carcinoma in situ (bottom) arising in
an adenomatous polyp. (d) Strong expression in colon carcinoma metastatic
to liver.
Source: Kolquist
et al
TELOMERASE MUTANTS?
- Telomerase has often been mutated in lab settings for studies on its structure and activity, but I have not come across mention of a "natural" mutation.
- A widely-publicized mutation was created in 2001, when UCSF scientists created a tiny mutation in the RNA template that caused the telomeres to be unstable after a few divisions. The mutation caused a dramatic halt in cancer cells' replication and division, in effect telling the cancer cells to commit suicide. As a potential "cure for cancer," it's still being researched in the hope that it will yield a treatment that doesn't damage normal human tissue the way other cancer treatments do (Trinkl, 2001).
REFERENCES
- Alberts, et al. Molecular Biology of the Cell. New York: Garland Science, 2002.
- Harrington, et al. A Mammalian Telomerase-Associated Protein. Science; January 1997 (275), 973-7). <http://www.sciencemag.org/cgi/content/full/275/5302/973?ijkey=MpBNqxqkLWxJA> Accessed 2003 March 12.
- Harrington, et al. Human telomerase contains evolutionarily conserved catalytic and structural subunits. Genes and Development; December 1997 (11), 3109-15. <http://www.genesdev.org/cgi/content/full/11/23/3109> Accessed 2003 March 12.
- Kech, Thomas. Chromosome Ends, Cancer, and Aging. <http://www.hhmi.org/annual98/research/chromo.html> Accessed 2003 March 12.
- Kilian, et al. Isolation of a candidate human telomerase catalytic subunit gene, which reveals complex splicing patterns in different cell types. Human Molecular Genetics; 1997 (12), 2011-9. <http://hmg.oupjournals.org/cgi/content/full/6/12/2011> Accessed 2003 March 12.
- Kolquist, et al. Expression of TERT in early premalignant lesions and a subset of cells in normal tissues. Nature; 1998 (19), 182-6. <http://www.nature.com/cgi-taf/DynaPage.taf?file=/ng/journal/v19/n2/full/ng0698_182.html#B7> Accessed 2003 March 12.
- Petty, Yvette. DNA Replication. <http://www.ncc.gmu.edu/dna/repanim.htm> Accessed 2003 March 13.
- Rfam 3.0. Washington Univ., St. Louis. <http://rfam.wustl.edu/index.html> Accessed 2003 March 13.
- Slish, Donald. Telomerase Demo. <http://faculty.plattsburgh.edu/donald.slish/Telomerase.html> Accessed 2003 March 12.
- Trinkl, Alice. UCSF Scientists Halt Tumor Growth by Manipulating Telomerase Enzyme. UCSF News Service, 2001.
Molecular
Biology Homepage
Sarah's
Molecular Homepage
Email Sarah