This web page was produced as an assignment for an undergraduate course at Davidson College.
Function and Structure of OmpF Porin
Introduction
The outer membrane protein F, better known as OmpF porin, is an integral membrane protein located in the outer membrane of the bacteria, Escherichia coli. OmpF porin is a non-specific transport channel that allows for the passive diffusion of small, polar molecules (600-700 Da in size) through the cell's outer membrane. Such molecules include water, ions, glucose, and other nutrients as well as waste products (Cowan et al., 1995). OmpF porins are found in a trimer formation within the outer cell membrane.
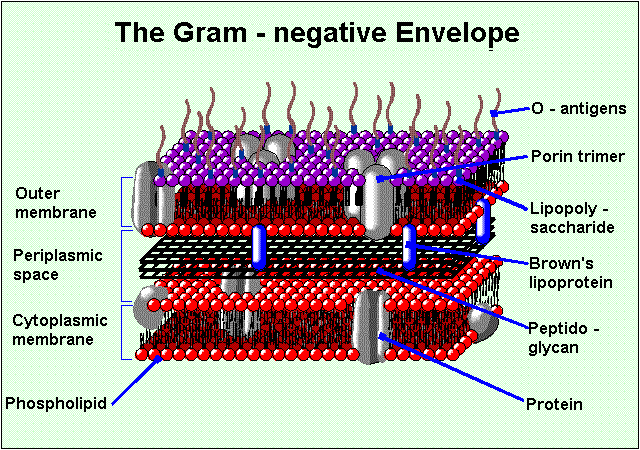
Figure 1. Diagram of the cell membrane. The cell membrane is comprised of two main structures: the internal membrane (cytoplasmic membrane) and the outer membrane. A thin peptidogylcan cell wall and a periplasmic space that contains substrate-binding enzymes separate the two membranes. The outer membrane contains groups of three porins called trimers. The OmpF porin is found in the trimer formation in E. Coli. Image produced by Dr. Henry Keil.
OmpF Porin Regulation
OmpF porin functions to regulate osmotic pressure between the cell and its surroundings. Many factors affect OmpF porin regulation, however, one of the more known is through the proteins EnvZ and OmpR. This process occurs in a series of steps:
1. A loss of osmotic pressure and an increase in solute concentration signals a disruption in the cell's homeostatic state. The structure of EnvZ is modified in response to this pressure change. EnvZ's changed structure allows it to add a phosphate group to itself using ATP.
2. EnvZ's phosphate group is subsequently transferred to and activates the OmpR protein.
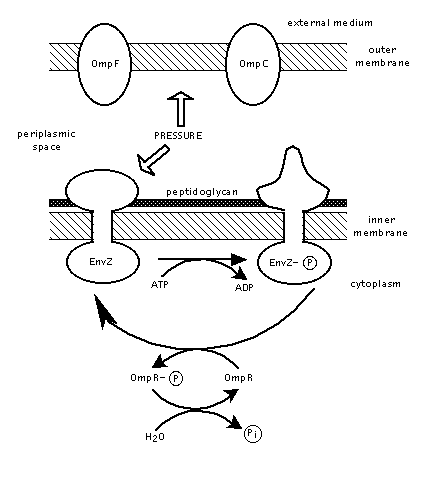
Figure 2. Diagram of EnvZ protein function as detailed in steps 1. and 2. Image produced by Dr. David Clark .
.
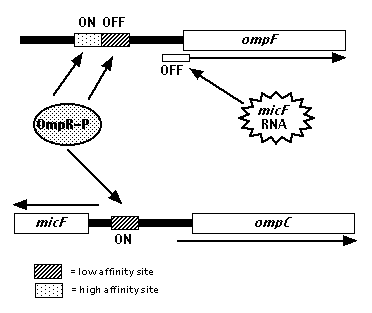
3. Because low osmolarity reduces the total amount of phosphorylated OmpR (OmpR-P) available in the cell, the OmpR-P that is produced attaches directly to the high affinity area of the OmpF promoter on the OmpF gene. Transcription of the gene is subsequently initiated.
4. The resulting OmpF mRNA undergoes translation and OmpF porin proteins are assembled and inserted into the outer membrane to prevent further osmotic pressure loss (Liu et al., 2001; Purves et al, 2001).
Figure 3. Regulation of OmpF gene transcription through OmpR-P protein as detailed in step 3. Phosphorylated OmpR can attach to either the OmpF gene promoter or the opposing OmpC promoter. When osmolarity is low, a lower amount of phosphorylated OmpR is available. The available OmpR-P therefore attaches to the high affinity promoter of the OmpF gene. If osmolarity is high, a greater number of OmpR-P is available. This allows the OmpR-P to attach to the lower affinity OmpC promoter region. Image produced by Dr. David Clark .
.
OmpF Porin Structure
--Primary Structure
The OmpF porin gene contains 1807 nucleotide bases. 1086 of these 1807 nucleotides make up the primary structure of the OmpF protein.
OmpF porin contains 340 amino acid residues. The figure below contains 22 extra amino acids (contained within the first 50 amino acids) that make up the amino terminus signal peptide. These 22 amino acids form a precursor sequence to the OmpF porin. They are eventually cleaved off to form OmpF porin (Inokuchi et al, 1982).
---------+----------+----------+----------+----------+
MMKRNILAVI VPALLVAGTA NAAEIYNKDG NKVDLYGKAV GLHYFSKGNG 50
ENSYGGNGDM TYARLGFKGE TQINSDLTGY GQWEYNFQGN NSEGADAQTG 100
NKTRLAFAGL KYADVGSFDY GRNYGVVYDA LGYTDMLPEF GGDTAYSDDF 150
FVGRVGGVAT YRNSNFFGLV DGLNFAVQYL GKNERDTARR SNGDGVGGSI 200
SYEYEGFGIV GAYGAADRTN LQEAQPLGNG KKAEQWATGL KYDANNIYLA 250
ANYGETRNAT PITNKFTNTS GFANKTQDVL LVAQYQFDFG LRPSIAYTKS 300
KAKDVEGIGD VDLVNYFEVG ATYYFNKNMS TYVDYIINQI DSDNKLGVGS 350
DDTVAVGIVY QF 362Figure 4. Amino acid structure of OmpF porin. This precursor form of OmpF porin contains 362 amino acids. The first 22 amino acids make up the amino terminus of this sequence. They are cleaved from the amino acid sequence as the precursor is being converted to OmpF porin (Inokuchi et al, 1982).
--Secondary Structure
The amino acid sequence provides the basis for the formation of the b-barrel channel of OmpF porin. Within its structure resides 3 a helix strands, 16 antiparallel b helix strands, and looping strands that connect the b helixes to each other (RCSB). Cartoon structures of OmpF porin have been deduced by x-ray crystallography. Click here for more information on x-ray crystallography.
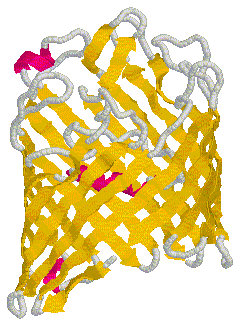
Figure 5. Secondary structure of OmpF porin monomer. The golden strands represent b helix strands. The pink strands represent a helix strands. The gray strands are loops that connect the many b helix strands to each other. Cartoon obtained from Determining physical properties of membranes.
A peculiarity of OmpF porin's structure is its narrow internal constriction channel located halfway into the protein. This narrow construction channel is lined with basic and acidic residues that allow the porin to perform its function and block both large molecules and solutes (Phale et al, 2001; Robertson et al, 2002). When three of these subunits attach to one another they form a trimer. OmpF porins are commonly found in the trimer formation within the outer membrane.
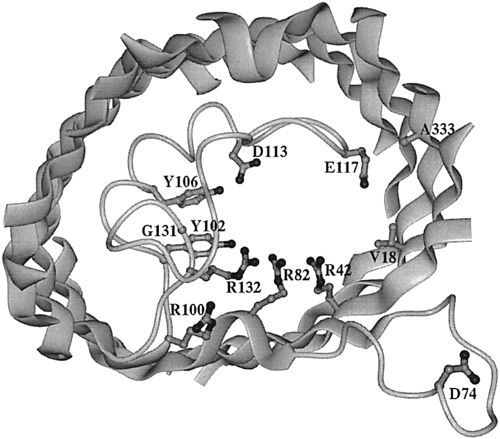 Figure 6. View of the extracellular side of an OmpF porin monomer channel. The interior of the protein is the channel constriction site. R42, R82, and R132 are the basic residues within the channel. Across from them reside D113 and E117, the acidic residues. The broad ribbons represent helix strands and the string represents the internal loop. The D74 loop attaches to other OmpF porin loops in order to form a trimer (Phale et al, 2001).
Figure 6. View of the extracellular side of an OmpF porin monomer channel. The interior of the protein is the channel constriction site. R42, R82, and R132 are the basic residues within the channel. Across from them reside D113 and E117, the acidic residues. The broad ribbons represent helix strands and the string represents the internal loop. The D74 loop attaches to other OmpF porin loops in order to form a trimer (Phale et al, 2001).
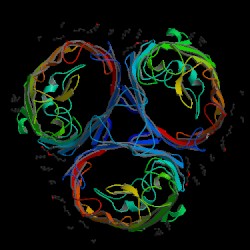
Figure 7. OmpF porin trimer. The looping strands of each porin protein bind with the adjacent loop of the next porin (Shown in the form of a triangle in the center of the three proteins). The constriction channels are the open spaces inside each porin. The internal loop can be seen as "wrapping around" this opening. Image produced by the Protein Data Bank.
References
Cowan SW, Garavito RM, Jansonius JN, Jenkins JA, Karlsson R, Konig N, Pai EF, Pauptit RA, Rizkallah PJ, Rosenbach JP. 1995 October. The structure of OmpF porin in a tetragonal crystal form [abstract]. Structure 3(10): 1041-1050. <http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Abstract&list_uids=8589999>. Accessed 2005 Feb 11.Purves W, Sadava D, Orians G, Heller H. Life: The Science of Biology. 6th Ed. U.S.A.: Sinauer Associates, Inc., 2001.
Liu X, Ferenci T. 2001. An analysis of multifactorial influences on the transcriptional control of ompF and ompC porin expression under nutrient limitation. Microbiology 147: 2981-2989. <http://mic.sgmjournals.org/cgi/content/full/147/11/2981>. Accessed 2005 Feb 13.
Inokuchi K, Mutoh N, Matsuyama S, Mizushima S. 1982. Primary structure of the OmpF gene that codes for a major outer membrane protein of Escherichia coli K-12. Nucleic Acids Research 10 (21): 6957-6968. <http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=326977>. Accessed 2005 Feb 12.
Phale S, Philippsen A, Widmer C, Phale V, Rosenbusch J, Schirmer T. 2001 May. Role of charged residues at the OmpF porin channel constriction probed by mutagenesis and simulation. Biochemistry 40 (21): 6319-6325. <http://pubs.acs.org/cgi-bin/article.cgi/bichaw/2001/40/i21/html/bi010046k.html>. Accessed 2005 Feb 15.
Robertson K, Tieleman D. 2002 September 25. Orientation and interactions of dipolar molecules during transport through OmpF porin. FEBS Letters 528(1-3): 53-57. <http://www.sciencedirect.com/science?_ob=ArticleURL&_udi=B6T36-46MC7MD-1&_coverDate=09%2F25%2F2002&_alid=246132751&_rdoc=1&_fmt=&_orig=search&_qd=1&_cdi=4938&_sort=d&view=c&_acct=C000058476&_version=1&_urlVersion=0&_userid=2665120&md5=6ae436a9ca7810e8330cd71e50617aa9>. 2005 Feb 15.
Research Collaboratory for Structural Bioinformatics (RCSB). The Protein Database. <http://www.rcsb.org/pdb/cgi/explore.cgi?job=chains&pdbId=2OMF&page=>. Accessed 2005 Feb 13.
Back to Davidson College Molecular Home
Any questions, comments, or suggestions?
E-mail pachampaloux@davidson.edu