This web page was produced as an assignment for an undergraduate course at Davidson College.
In literature, this protein has been alternatively referred to as: Amyloid Protein, Amyloid Protein Peptide, ß-Amyloid Precursor Protein, Amyloid ß-Precursor Protein, and other variations of these combinations .
Introduction
Human Amyloid Precursor Protein (abbreviation: APP) is the protein product of a single gene located on chromosome 21. This transmembrane protein is expressed in many different types of cells, especially in fetal brain, heart, spleen, and kidney tissues (Abcam, 2005). It is most abundantly expressed in the anterior perisylvian cortex-opercular gyri and in the frontal lobe of the cortex of the adult brain (Abcam, 2005). Scientists have identified ten isoforms of APP, which are created by alternative splicing (Upstate, 2005). The three most common isoforms are termed C, B, and A. They have 695, 751, and 770 amino acids, respectively. The C isoform (695 amino acids) is primarily present in neurons, while the B isoform (751 amino acids) isoform predominates T-lymphocytes (Abcam, 2005).
Structure
Primary Structure: Amino Acid Sequence
The three most common APP isoforms have different amino acid (abbreviation: a.a.) sequences due to alternative splicing. The isoforms B and A have an insertion comprised of their amino acids 288-345 that make them longer than isoform C. Isoform A has an insertion comprised of its amino acids 345-364 that makes it longer than both isoforms B and C.
The amino acid sequence of the 695 a.a. C isoform is as follows (courtesy of the NCBI):
1 mlpglallll aawtaralev ptdgnaglla epqiamfcgr lnmhmnvqng kwdsdpsgtk
61 tcidtkegil qycqevypel qitnvveanq pvtiqnwckr grkqckthph fvipyrclvg
121 efvsdallvp dkckflhqer mdvcethlhw htvaketcse kstnlhdygm llpcgidkfr
181 gvefvccpla eesdnvdsad aeeddsdvww ggadtdyadg sedkvvevae eeevaeveee
241 eadddedded gdeveeeaee pyeeatertt siattttttt esveevvrvp ttaastpdav
301 dkyletpgde nehahfqkak erleakhrer msqvmrewee aerqaknlpk adkkaviqhf
361 qekvesleqe aanerqqlve thmarveaml ndrrrlalen yitalqavpp rprhvfnmlk
421 kyvraeqkdr qhtlkhfehv rmvdpkkaaq irsqvmthlr viyermnqsl sllynvpava
481 eeiqdevdel lqkeqnysdd vlanmisepr isygndalmp sltetkttve llpvngefsl
541 ddlqpwhsfg adsvpanten evepvdarpa adrglttrpg sgltniktee isevkmdaef
601 rhdsgyevhh qklvffaedv gsnkgaiigl mvggvviatv ivitlvmlkk kqytsihhgv
661 vevdaavtpe erhlskmqqn gyenptykff eqmqn
The amino acid sequence of the 751 a.a. B isoform is as follows (courtesy of the NCBI):
1 mlpglallll aawtaralev ptdgnaglla epqiamfcgr lnmhmnvqng kwdsdpsgtk
61 tcidtkegil qycqevypel qitnvveanq pvtiqnwckr grkqckthph fvipyrclvg
121 efvsdallvp dkckflhqer mdvcethlhw htvaketcse kstnlhdygm llpcgidkfr
181 gvefvccpla eesdnvdsad aeeddsdvww ggadtdyadg sedkvvevae eeevaeveee
241 eadddedded gdeveeeaee pyeeatertt siattttttt esveevvrev cseqaetgpc
301 ramisrwyfd vtegkcapff yggcggnrnn fdteeycmav cgsaipttaa stpdavdkyl
361 etpgdeneha hfqkakerle akhrermsqv mreweeaerq aknlpkadkk aviqhfqekv
421 esleqeaane rqqlvethma rveamlndrr rlalenyita lqavpprprh vfnmlkkyvr
481 aeqkdrqhtl khfehvrmvd pkkaaqirsq vmthlrviye rmnqslslly nvpavaeeiq
541 devdellqke qnysddvlan miseprisyg ndalmpslte tkttvellpv ngefslddlq
601 pwhsfgadsv pantenevep vdarpaadrg lttrpgsglt nikteeisev kmdaefrhds
661 gyevhhqklv ffaedvgsnk gaiiglmvgg vviatvivit lvmlkkkqyt sihhgvvevd
721 aavtpeerhl skmqqngyen ptykffeqmq n
The amino acid sequence of the 771 a.a. A isoform is as follows (courtesy of the NCBI):
1 mlpglallll aawtaralev ptdgnaglla epqiamfcgr lnmhmnvqng kwdsdpsgtk
61 tcidtkegil qycqevypel qitnvveanq pvtiqnwckr grkqckthph fvipyrclvg
121 efvsdallvp dkckflhqer mdvcethlhw htvaketcse kstnlhdygm llpcgidkfr
181 gvefvccpla eesdnvdsad aeeddsdvww ggadtdyadg sedkvvevae eeevaeveee
241 eadddedded gdeveeeaee pyeeatertt siattttttt esveevvrev cseqaetgpc
301 ramisrwyfd vtegkcapff yggcggnrnn fdteeycmav cgsamsqsll kttqeplard
361 pvklpttaas tpdavdkyle tpgdenehah fqkakerlea khrermsqvm reweeaerqa
421 knlpkadkka viqhfqekve sleqeaaner qqlvethmar veamlndrrr lalenyital
481 qavpprprhv fnmlkkyvra eqkdrqhtlk hfehvrmvdp kkaaqirsqv mthlrviyer
541 mnqslsllyn vpavaeeiqd evdellqkeq nysddvlanm iseprisygn dalmpsltet
601 kttvellpvn gefslddlqp whsfgadsvp antenevepv darpaadrgl ttrpgsgltn
661 ikteeisevk mdaefrhdsg yevhhqklvf faedvgsnkg aiiglmvggv viatvivitl
721 vmlkkkqyts ihhgvvevda avtpeerhls kmqqngyenp tykffeqmqn
Secondary And Tertiary Structures
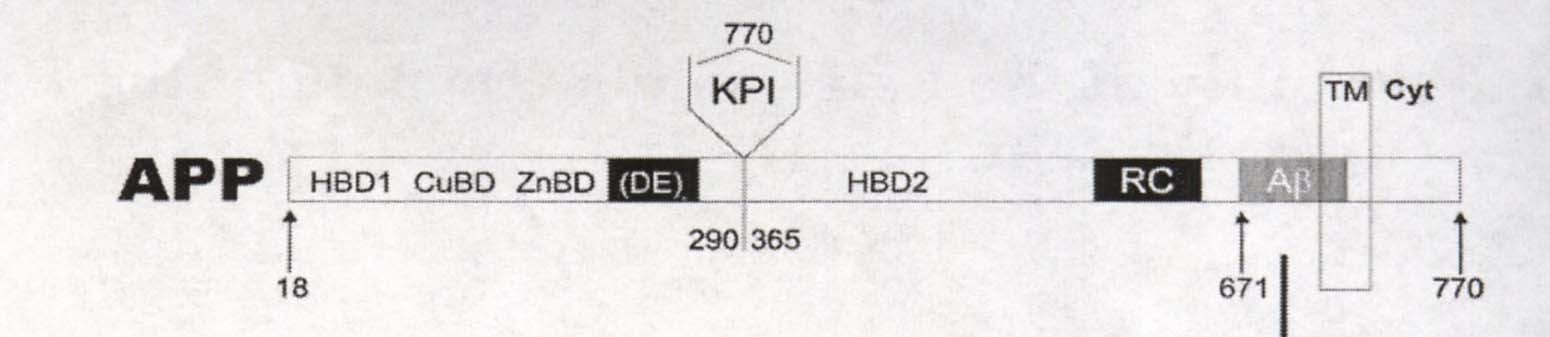
The secondary and tertiary structures of the Amyloid Precursor Protein are very complicated because of the presence of multiple distinct domains (subsections of a protein). The majority of the protein's domains are extracellular. Beginning from the N-terminus and traveling down towards the C-terminus, there is a heparin-binding/growth-factor-like domain (HBD1), a copper binding domain (CuBD), a zinc binding domain (ZnBD), an acidic Asparatic Acid and Glutamic Acid portion (DE), a Kunitz-type protease inhibitor domain (KPI) (except in isoform C), another heparin binding domain (HBD2), a random coil region (RC), and a 28 amino acid amyloid beta region (Aß) in the extracellular portion of the protein. In some literature, a region containing both parts of HBD2 and the RC region is termed E2 domain (Wang Y, Ha Y, 2004). A transmembrane portion of the protein follows, containing about 12 amino acids of the amyloid beta region. Finally the remainder of the protein is a cytoplasmic tail region. See Figure 1 below for a visual representation of the general structure of APP.

Figure 1. Cartoon of Amyloid Precursor Protein Domain Sequence. Numbers represent amino acids. E2 domain is not labeled. Image courtesy of: (Botelho MG, et al, 2003), permission granted by Dr. Sergio T. Ferreira: 2/16/05.
Domain Structures
The secondary and tertiary structures of several domains of the Amyloid Precursor Protein have been studied and described in detail.
· N-Terminal Heparin-Binding/ Growth Factor-like Domain (HBD1) structure
The Growth-Factor-like domain of APP has been described to 1.8Å (Rossjohn J, et al., 1999). It occurs in all isoforms between amino acids 28 through 123 of the APP protein. This domain contains one alpha-helix comprised of 13 amino acids and nine ß-sheets that are mostly comprised of very short strands of three amino acids. There are three disulfide bridges within this domain that hold it into a "compact, globular" shape. One side of the domain has an extremely positively charged surface due to positively charged amino acids such as K and R. Furthermore, there is a non-polar or hydrophobic region directly next to the positive portion of the domain. Amino acids such as F, P, V, W, and M dominate this non-polar region (Rossjohn J, et al., 1999).
· Copper Binding Domain (CuBD) structure
The Copper Binding Domain has been studied with triple resonance multidimensional NMR spectroscopy (Barnham KJ, et al., 2003). It occurs in all isoforms between amino acids 124-189 in all isoforms. This domain contains one alpha-helix comprised of 12 amino acids and three ß-sheets with 5, 6, or 7 amino acids each. There are three disulfide bridges in the Copper Binding Domain. The surface of this domain has both extremely positive and negative portions. The negatively charged areas contain amino acids D and E, while the positively charged contain the amino acids K and H. Finally, there is a small non-polar or hydrophobic central region of the Copper Binding Domain, comprised of a combination of amino acids A, W, V, M, and L (Barnham KJ, et al., 2003). See Figure 2 below for a visual representation of this domain.
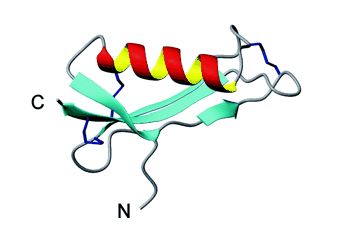
Figure 2. Ribbon cartoon of CuBD domain of APP. Red and yellow helix is the alpha-portion, blue sheets are the ß-portions. Image courtesy of (Barnham KJ, et al., 2003), permission granted by Dr. Michael Parker: 2/24/05.
· E2 Region (Containing portions of HBD2 and RC region) structure
The E2 region contains portions of the second Heparin-Binding Domain and the randomly coiled region. It has been described to 2.8 Å (Wang Y, Ha Y, 2004). This domain consists of amino acids 355-546 of APP isoform B, at amino acids 373-526 of isoform A, and at amino acids 298-470 of isoform C. The E2 region has six total alpha-helices, termed A-F. These helices form two "coiled coil" substructures (meaning parts are folded over each other). B-helix and part of C-helix form the N-terminal antiparallel, two-stranded coiled substructure, while the rest of C, D, E, and F-helices form the antiparallel, triple-stranded coiled substructure of the C-terminal. These substructures are considered "antiparallel" because in folding the N-terminal of one helix is aligned the C-terminal of the adjacent helices (i.e. the N-terminal of B is aligned with the C-Terminal of A and C). See Figure 3 below for a visual representation of this folding.
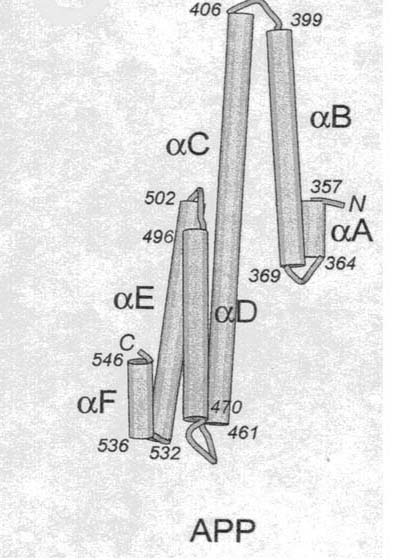
Figure 3. Cartoon of E2 region of APP. Numbers represent amino acids. N is the abbreviation for N-Terminal, C is the abbreviation for C-Terminal. Image courtesy of (Wang Y, Ha Y, 2004), permission granted by Dr. Ya Ha: 2/16/05.
There are two glycosylation sites within E2. They occur at amino acid Asparagine at positions 523 and 552. Researchers also found that one E2 region of one APP can bind with the E2 region of another APP to form an antiparallel dimer in solution. It is the only APP domain that forms a dimer in solution. Positively charged amino acids line the outside surface of this dimer (Wang Y, Ha Y, 2004).
Function
The overall physiological function of Amyloid Precursor Protein has not yet been determined definitely. Scientists have postulated that transmembrane APP serves as a cell surface receptor , as a heparin binding site, as a precursor to a growth-factor-like agent (Wang Y, Ha Y, 2004) , and as a "regulator of neuronal copper homeostasis" (Barnham KJ, et al., 2003). These hypotheses appear valid because the structures of specific domains of APP possess specific characteristics that are indicative of these functions.
· N-Terminal Heparin-Binding/ Growth Factor-like Domain (HBD1) proposed function
The secondary structure of HBD1 domain mostly consists of ß-sheets, like known cystein-rich growth factors (Rossjohn J, et al., 1999). The disulfide bonds in HBD1 also form ß-loops between ß-sheets, which are particularly indicative of growth factors. Because of these similarities and because HBD1's amino acid sequence is consistent with those of other known growth factors, HBD1 appears to function as growth-factor. Furthermore, the HBD1 region possesses an extremely positively charged surface, which would allow the anion (negatively charged) heparin, to bind. Thus it likely functions as a heparin binding site as well (Rossjohn J, et al., 1999).
· Copper Binding Domain (CuBD) proposed function
CuBD contains negatively charged Histadine amino acids, which are necessary for copper binding. Copper is a positively charged cation that is attracted by negative charges. Furthermore, the single alpha-helix and triple ß-sheet combination in CuBD is also present in three proteins with known copper chaperone activity (transport). Thus, CuBD has properties indicative of and similar to other copper binding protein . This region likely serves as a regulator of copper concentration within the body (Barnham KJ, et al., 2003).
· E2 Region (Containing portions of HBD2 and RC region) proposed function
Because of the E2 folding structure, it can form an antiparallel dimer. Theoretically, antiparallel dimers could form between two E2 regions of two APPs embedded in the same cell membrane, or between two APPs embedded in separate cell membranes. Two cells could communicate with or adhere to each other via E2 region dimer formation. Thus, APP proteins have the potential to function in cell signaling and adhesion processes (Wang Y, Ha Y, 2004).
Dysfunction and Alzheimer’s Disease
Particular proteases termed alpha-, ß-, and gamma-secretase can cleave Amyloid Precursor Protein into smaller protein fragments. Scientists hypothesize that this fragmentation of APP allows many of the functions proposed above to be fulfilled as part of normal brain processes. In other words, some APP domains need to be separated from each other to function as proteins in their own right. One particular pathway of ß-secretase mediated cleavage of APP followed by gamma-secretase mediated cleavage, results in the formation of Amyloid-ß fragment (made of 40, 42, or 43 amino acids). See the right portion of Figure 4 below for a visual representation of this splicing.
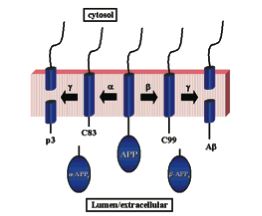
Figure 4. Cartoon of Two Alternative Pathways of Secretase Mediated Cleavage of APP. In one instance, C83 and a extracellular soluble fragment called alpha- APPs results from alpha-secretase cleavage of APP. Further cleavage of C83 by gamma-secretase forms p3. In the other case, ß-secretase cleaves APP to produce extracellular soluble ß-APPs and C99. Further cleavage of C99 by gamma-secretase, produces the suspicious Amyloid-ß protein fragment. Image courtesy of (Wolfe MS, 2001), permission granted by Dr. Michael Wolfe: 2/16/05.
Scientists hypothesize that in moderate amounts, the Amyloid-ß fragment may serve some useful purpose. In excess, however, particularly sticky Amyloid-ß fragments aggregate and eventually become "senile plaques" (Selkoe DJ, 1991). These plaques have been linked to the destruction of neurons. subsequent loss of memory and cognitive function, and the development of Alzheimer's Disease. Researchers are currently working to better understand the normal physiological function of APP and the Amyloid-ß fragment. The future development of secretase blockers or anti-ß drugs could lead to prevention or reversal of to Alzheimer's disease (Alzheimer's Association, 2004). Caution must be employed, however, because so as not to impair normal brain function.
For more information about Alzheimer's Disease and the "amyloid hypothesis", visit the following web sites:
The Alzheimer's Association: http://www.alz.org
The Alzheimer's Disease Education and Referral Center: http://www.alzheimers.org/
References and Works Consulted:
Abcam. Product Datasheet for ab7875. <http://www.abcam.com/framework/popups/popup_printable_ds.cfm?intAbID=7875>. Accessed 2005 Feb 8.
Alzheimer’s Association. 2004 Dec 10. Fact Sheet: About Alzheimer drugs targeting beta-amyloid and the “amyloid hypothesis.” <http://www.alz.org/Resources/TopicIndex/BasicFacts.asp#causes>. Accessed 2005 Feb 8.
Barnham KJ, et al. 2003 May 9. Structure of the Alzheimer’s Disease Amyloid Precursor Protein Copper Binding Domain. J Biological Chemistry 278 (19): 17401-17407.
Betelho MG, et al. 2003 Sept 5. Folding and Stability of the Extracellular Domain of the Human Amyloid Precursor Protein. J Biological Chemistry 278 (36): 34259-34267.
Rossjohn J, et al. 1999 Apr. Crystal structure of the N-terminal, growth factor-like domain of Alzheimer amyloid precursor protein. Nature Structural Biology 6 (4): 327-331.
Selkoe DJ. 1991 Nov. Amyloid Protein and Alzheimer’s Disease. Scientific American 265 (5): 68-78.
Upstate. Amyloid Precursor Protein and Amyloid Beta. <http://www.upstate.com/features/app_lp.asp?c=221&r=556>. Accessed 2005 Feb 8.
Wang Y, Ha Y. 2004 Aug 13. The X-Ray Structure of an Antiparallel Dimer of the Human Amyloid Precursor Protein E2 Domain. Molecular Cell 15: 343-353.
Wolfe MS. 2001 Jun 21. Secretase Targets for Alzheimer’s Disease: Identification and Therapeutic Potential. J Medicinal Chemistry 44(13): 2039-2060.