* This webpage was created as an undergraduate assignment at Davidson College, Davidson, NC*
Orthologs are proteins that have similar conserved sequences among species. Orthologs can provide insight into the evolutionary histories and functions of a particular protein. The telomerase of Tribolium castaneum was the first the high resolution protein structure determined (Gillis et al. 2008). Its sequence can be compared to other species to analyze the difference in function across phylogenetic groups and can provide insight into the origin of telomerase.
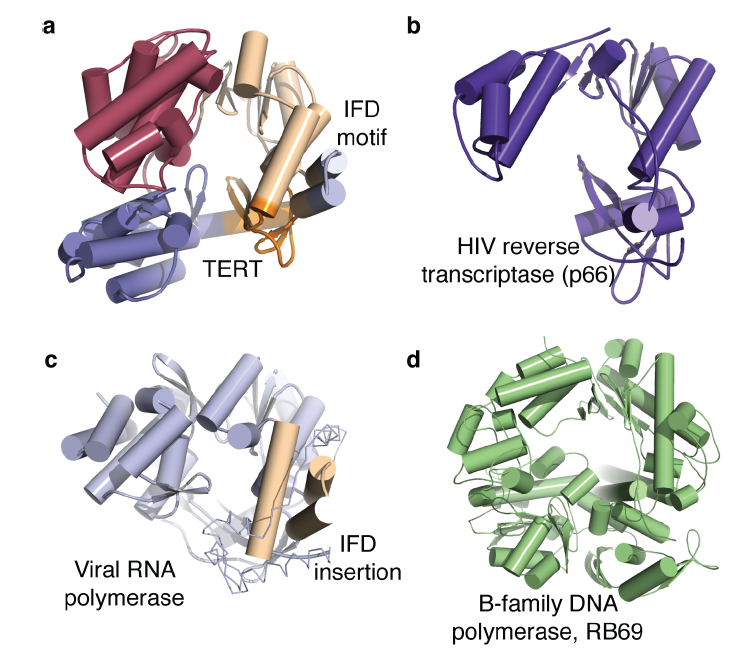
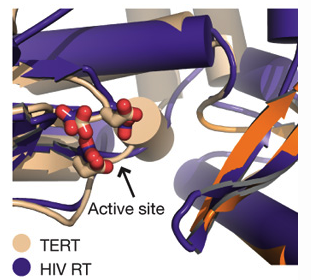
Telomerase is specific to eukaryotic organisms since the end replication problem applies only to linear DNA. However, Gillis et al. describes telomerase as part of the retroviral reverse transcriptase superfamily (2008). The researchers compare the telomerase of T. castaneum to the closest homologue of telomerase, HIV reverse transcriptase, to demonstrate a high degree of similarity between the active sites of the two proteins (Figure 2). In addition, the ringlike structure of telomerase is similar to the structure of viral RNA polymerases, B-family DNA polymerases and the HIV reverse transcriptase (p66) (Figure 1). The similar structure implies a similar function, which is supported by the fact that they all add nucleotides to nucleic acid strands. This similarity demonstrated in structure and function among these different proteins suggests a close evolutionary relationship (Figure 5). However, since these comparisons are between homologues rather than orthologs, the following discussion will focus on the orthologs of telomerase.


Figure 2 (right). Superposition of HIV RT (tan ) and TERT (purple) active sites shows a high similarity in structure, which will hypothetically confer a similarity in function.
©2007 Gillis et al. Permission Pending.
The motifs of the TERT subunit of telomerase is conserved across eukaryotic species. Gillis et al. state that TER, the RNA template subunit, varies in size, sequence and structure among different phylogenetic groups but the core structural elements are conserved among species (2008). We will focus on TERT because it is the catalytic subunit of telomerase. The 7 motifs that are associated with reverse transcriptase are conserved throughout the different species analyzed in Bryan et al. (1998). These motifs are in the 'fingers' and 'palm' domain and include motifs 1, 2, A, B', C, D and E (Figure 3A). The T motif is specific to telomerase and is also conserved across these species. Another difference between telomerase and retroviral or retrotransposon reverse transcriptase is the Arg residue in motif 1 and the aromatic Tyr or Phe after 2 Asp residues in motif C (Figure 3b). In addition, a CP motif appeared only for the ciliates, as shown in Figure 3. The paper posits that the CP motifs are specific to the ciliates, which may help to explain the enormous increase in telomere numbers found in ciliates as compared to other eukaryotes.
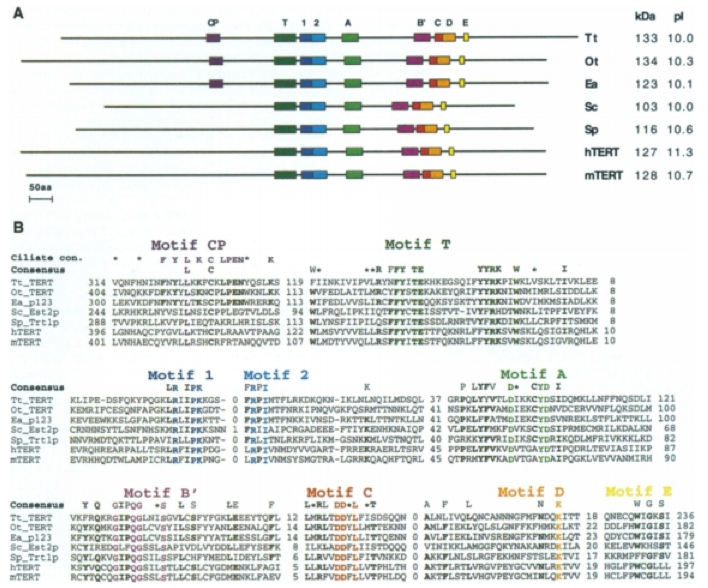
Figure 3. (A) Motifs conserved among 7 species. Motifs 1, 2, A, B', C, D and E are associated with reverse transcriptase. The T motif is specific to telomerase and the CP motif is specific to the ciliates, which have a higher telomere number than the remaining species. (B) Comparison of amino acid sequences across the 7 species (hTERT = human, mTERT = Mus muscularus, Sc.Est.2p= Saccharomyces cerevisiae, Sp. trt1p = schizosaccharomyces pombe, Tt TERT=tetrahymena thermophila, Ot. Tert =oxytricha trifallas, Ea. p123=euplotes aediculatus). The colored amino acids indicate the residues that are conserved within the labeled motifs. The consensus sequence indicates residues that are conserved in more than 5 of the species, which are shown in bold. Lastly, the ciliate consensus sequence only includes the conserved sequences in ciliates that correspond to the CP motif.
©1998 Bryan et al. Permission Pending.
Gillis et al. found a similar pattern in conserved sequences among various phylogenetic groups. First, they compared the motifs of T. castaneum to those of yeast, human and T. thermophila. The yeast, human and T. thermophila had all the motifs present in the T. castaneum but had additional sequences (GQ) upstream of the CP motif (Figure 4). One motif identified by this group not mentioned by the previous group is the IFD motif. The three aspartic acids at the active site that are part of motifs A and C are necessary for TERT functioning because replacement with alanine results in complete TERT inactivity. One difference is the IFD motif insertion between motifs A and B’ of TERT that is necessary for telomerase processivity. CTE or thumb domain is not found in any other known protein but structurally it is most similar to the thumb domains of the other RT and polymerases.
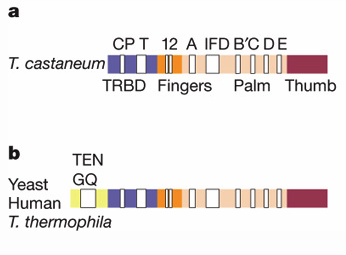
Figure 4. General motif comparison between T. castaneum (a) and yeast, human and T. thermophila (b). The white boxes indicate the labeled motifs and the colors indicate the labeled domains.
© 2008 Gillis et al. Permission Pending.
Gillis et al. compares the seqeunces of 7 different species to T. castaneum and finds striking similarity. The motifs are clearly conserved throughout the species, with minor residue alterations that do not greatly change the structure of the protein due to their similarity in chemical properties (Figure 5). The important secondary structures are also highlighted. Alpha 10 interacts with TBD and a deletion or mutation of this motif disrupts TERT activity. The lysine at K210 within the alpha 10 structure is important because it will interact with the negatively charged DNA backbone. The mouse and human sequences have a substitution of asparagine at this position, the yeast has a glutamate, the T. thermophila has an arginine and the E. aediculatus has a threonine, all of which carry polar side chains. Similarly, K406, K416, K418 and N423 all carry polar side chains to interact with the DNA backbone. A deletion or mutation of alpha 19 (minor groove) in yeast and human TERT causes loss of TERT processivity. Finally, Gillis et al. emphasizes the importance of the beta hairpain in the T motif, which is mostly conserved throughout all species. The beta hairpin in T motif interacts with motifs 1 and 2, which places the opening of the TRBD pocket in the interior of the ring close to the active site. The beta hairpin is posited to be an allosteric effector switch to couple RNA binding inside the ring to placement of the RNA template at the active site.

Figure 5. A Clustal W2 comparison of TERT sequences across a range of phylogenetic groups showing both conserved motifs (colored boxes) and secondary structures (beta strands as arrows and alpha helices as cylinders) as described in Telomerase Structure and Function. The K210 (brown) and K406, K416, K418 and N423 (yellow) residues are conserved polar residues to interact with the negatively charged DNA backbone. The sequences are labeled as follows: TRICA = Tribolium castaneum, MOUSE = Mus musculus, HUMAN = Homo sapiens, ARATH = Arabidopsis thaliana, YEAST = Saccharomyces cerevisiae,SCHPO = Schizosaccharomyces pombe, TETTH = Tetrahymena thermophila, and EUPAE =Euplotes aediculatus.
©2007 Gillis et al. Permission Pending.
A comparison of multiple species confirms the phylogenetic relationships of these species and indicates an ancient origin of telomerase. The relationships of these organisms follow what you would expect given current knowledge of phylogenies. Notably, the sequences of HIV reverse transcriptase, DNA polymerase and Hepatitis C are different from telomerase in all species, which indicates that the telomerase of all species is more similar to one another than to the other polymerases, supporting the hypothesis that they all evolved from one ancient origin (Figure 6).
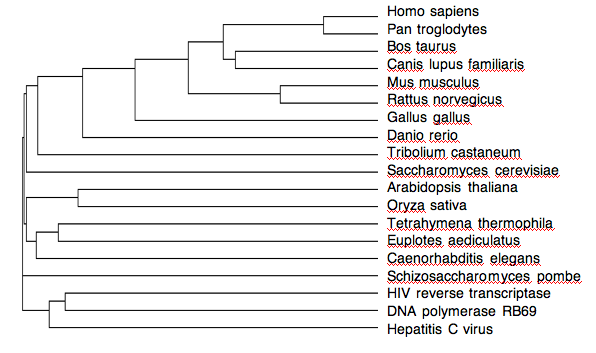
Figure 6. Phylogeny of different species using telomerase or homologs of telomerase (Clustal W2)
©2010 Danielle Jordan. Phylogeny created by the program Clustal W2 with sequences publicly available at NCBI and labeled by Danielle Jordan.
References
[1] Gillis AJ, Schuller AP, Skordalakes E. Structure of the Tribolium castaneum telomerase catalytic subunit TERT. Nature 2008; 455(7213):633-7. Pubmed
[2] Bryan TM, Sperger JM, Chapman KB, Cech TR. Telomerase reverse transcriptase genes identified in tetrahymena thermophila and oxytricha trifallax. Proc Natl Acad Sci U S A 1998 Jul. 21;95(15):8479-85. Article
©2010 Danielle Jordan, Biology Department, Davidson College, Davidson, NC
Questions? Comments? Email me at dajordan@davidson.edu