*This website was produced as an assignment for an undergratuate course at Davidson College.*
My Favorite Protein:
HIV Reverse Transcriptase
View Structure of HIV-1 Reverse Transcriptase in Jmol (HIV-1 Reverse Transcriptase)
Brief Introduction:
HIV-1 reverse transcriptase is the enzyme responsible for creating a DNA copy of the HIV viral genome. It uses the viral genome (which is composed of single stranded RNA) as a template to create a double stranded DNA copy of the viral genome so that it can be incorporated into the host cell and use the host cell’s machinery to make many copies of itself (Sadava et al., 2008). This enzyme has DNA polymerase activity for creating a DNA copy of the viral genome and ribonuclease H (RNase H) activity for cleaving the viral template ssRNA after it has been copied (Rodgers et al., 1995).
Structure and Function:
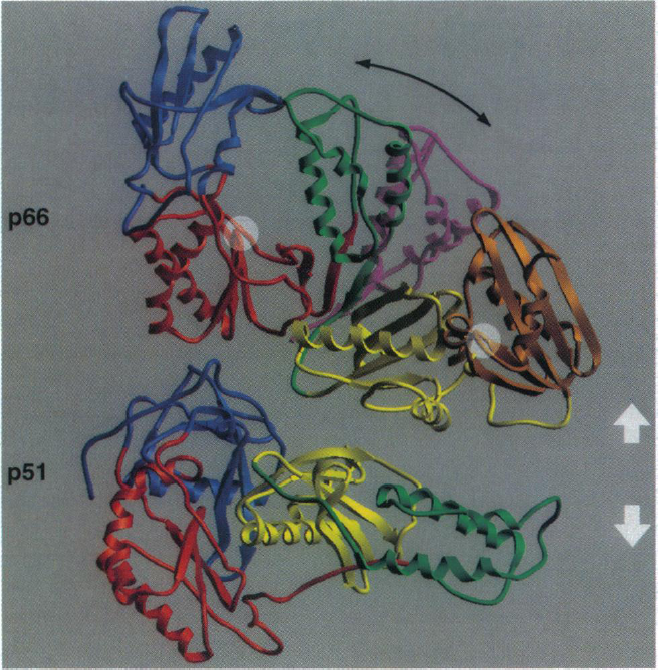
HIV-1 reverse transcriptase is composed of two primary subunits: p66 (66-kDa) and p51 (51-kDa). The p51 subunit is created by a post-translational modification where the C-terminus of the protein (p66/p66 homodimer) is cleaved (Rodgers et al., 1995). The p66 subunit is composed of 560 amino acids and the p51 subunit is composed of the first 450 amino acids of the p66 subunit (Shafter et al., 1999). Both of the p66 and p51 subunits have several similar structural motifs (Figure 1). The most highly conserved portion of this protein is the core region which includes the p66 connection domain and the p51 fingers, thumb and connection domains. These domains are so highly conserved because they are important for interactions between the p51 and p66 subunits, which are crucial for maintaining the structure of the heterodimer. Although both of the subunits have corresponding structural motifs, these motifs are arranged very differently in each of the subunits (Figure 1). This is because each of the subunits has distinct and unique functions. The p66 subunit is the catalytic subunit and has DNA polymerase and RNase H activity. The p51 subunit is responsible for stabilizing the p66 subunit.
The fact that this enzyme is a heterodimer is very important because it allows for a high degree of flexibility within this protein, which is critical for proper enzymatic function. During the enzymatic activity of this protein, HIV-1 reverse transcriptase goes through three significant conformational changes: 1) when binding to the nucleic acids, 2) after binding to the nucleic acids, before catalytic activity and 3) after catalytic activity in order to allow for movement of the enzyme along the template strand to catalyze the next nucleotide addition (Patel et al., 1995). This sequence of events shows that flexibility within this enzyme is very important and both the p66 and p51 domains are important for facilitating these movements.

Image from Rodgers et al. 1995. Permission pending.
Figure 1. This figure depicts the basic structure of the HIV reverse transcriptase with different domains highlighted (blue: fingers, red: palm, green: thumb, yellow: connection and brown: RNase H). The purple domain indicates the change in shape of the p66 domain when the template strand is bound to this enzyme. The p66 and p51 subunits are indicated and the white arrows show that these two domains have been separated. The white circles highlight the location of the polymerase and RNase active sites.
p66 subunit:
The p66 subunit assumes an active and inactive conformation. In the inactive conformation, the thumb domain of the p66 subunit lies over the active site and interacts with the p66 fingers domain (Figure 1). In the active conformation, the p66 thumb domain moves away from the p66 fingers domain and exposes the catalytic site (Figure 1). The catalytic site is located in the palm sub-domain of the p66 subunit (Figure 1) and the amino acid sequence at this region (YMDD) is highly conserved among retroviruses (Patel et al., 1995). When the p66 domain changes shape in order to allow interaction with RNA, the change in the shape of active site in p66 occurs in such a manner that only single stranded RNA can interact with the protein (Patel et al., 1995). This is important because this enzyme’s job is to act on a single stranded RNA and make a DNA copy of this template.
There are several regions of the p66 subunit that are critical for proper enzymatic function. The aspartic acid side chains located in the active site of the p66 subunit (Asp110, Asp185, and Asp186), are involved in interacting with the phosphate groups on nucleotide triphosphates that are added to the growing DNA strand, and are involved in breaking the bonds between these phosphate groups, which is crucial for powering the process of DNA polymerization (Figure 2; Patel et al., 1995). The tyrosine at amino acid residue 115 is specifically responsible for interacting with the nucleotide itself that is being added to the DNA strand being synthesized (Figure 2). The hydrophobic interactions between this amino acid residue and the incoming nucleotide are important for proper synthesis of the DNA strand (Patel et al., 1995). It is believed that this amino acid residue is analogous to tyrosine residues found in other polymerase enzymes (such as the mammalian polymerase α), which are important in making sure that the proper complementary base is being added to the strand that is being polymerized (Patel et al., 1995). Other amino acid residues (specifically Gln151-Trp153, Leu74-Arg78 and Glu89) near the active site located in the thumb and fingers domain are responsible for holding the RNA template strand in place (Patel et al., 1995). Once a single-stranded DNA copy of the viral genome has been made, the reverse transcriptase catalyzes the formation of double-stranded DNA in the p66 active site.
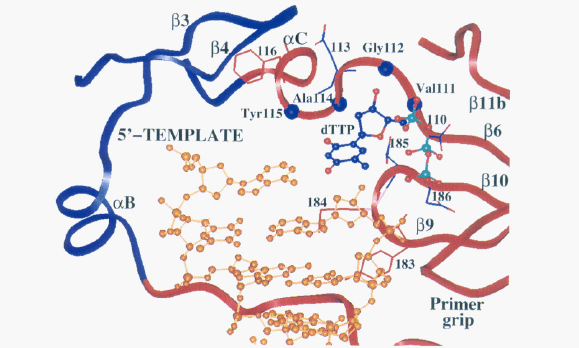
Image from Patel et al., 2005. Permission pending.
Figure 2. This ribbon diagram depicts the active site of HIV-1 reverse transcriptase. The palm region is colored in red, the fingers region is colored in blue and the DNA is colored gold. The side chains and amino acid residues that are important for interacting with the template strand and incoming nucleotide are highlighted. In addition, the different regions of secondary structure (beta pleated sheets and alpha helices) that contain these amino acid residues are also depicted. This view of the active site gives a clear indication of how the structure of the active site allows for very specific interactions with different regions of the incoming nucleotide and the RNA template strand.
p51 subunit:
Although all of the enzymatic activity is localized in the p66 domain, the p51 domain plays an important role in the function of this enzyme. It is very important for stabilizing the p66 subunit (Figure 3), which is necessary for the catalytic subunit to do its job properly (Shafer et al., 1999). The connection domain of the p51 subunit is important for tRNA binding (tRNA functions as the replication primer for HIV-1 reverse transcriptase; Jacques et al., 1994). tRNAs tend to bind with higher affinity to heterodimers rather than homodimers (Jacobo-Molina and Arnold, 1991), thus the cleavage of the p66/p66 homodimer into the p66/p51 heterodimer is important for more efficient enzymatic function. More recently scientists have found that deletions at particular amino acid sites within the p51 domain affect the ability of this protein to form a heterodimer and decrease the polymerase activity of the enzyme (Upadhyay et al., 2010). Thus, although this subunit is not directly involved in the enzymatic activity of this enzyme, it is clear that it is very important for proper functioning of the enzyme.
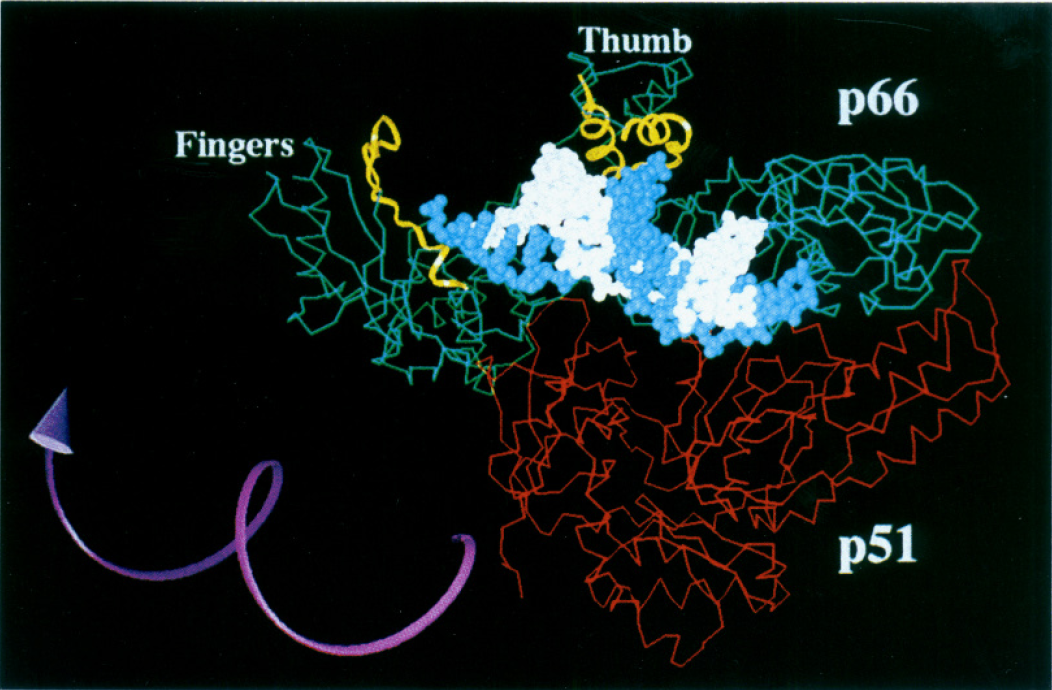
Image from Patel et al. 1995. Permission pending.
Figure 3. HIV-1 reverse transcriptase active site. The p66, p51, p66-thumb and p66-fingers domains are all labeled on the diagram. The white strand indicates the primer and the template strand is in blue. The amino acid residues that are thought to interact with the DNA are highlighted in yellow. The purple arrow shows the direction the enzyme turns in order to catalyze the polymerization of the growing DNA strand. This image also clearly indicates the importance of the p51 in stabilizing the heterodimer.
Works Cited:
Jacobo-Molina A and Arnold E. 1991. HIV reverse transcriptase structure-function relationship. Biochemistry 30: 6351-6361.
Jacques PS, Birgitta MW, Howard KJ and Le Grice SFJ. 1994. Modulation of HIV-1 reverse transcription function in “selectively deleted” p66/p51 heterodimers. Journal of Biological Chemistry 269: 1388-1393.
Patel PH, Jacobo-Molina, Ding J, Tantillo C, Clark AD, Raag R, Nanni RG, Hughes SH and Arnold E. 1995. Insights into DNA polymerization mechanisms from structure and function analysis of HIV-1 reverse transcriptase. Biochemistry 34: 5351-5363.
Rodgers DW, Gamblin SJ, Harris BA, Ray S, Culp JS, Hellmig B, Woolf DJ, Debouck C and Harrison SC. 1995. The structure of unliganded reverse transcriptase from the human immunodeficiency virus type 1. Proceedings of the National Academy of Science 92: 1222-1226.
Sadava D, Heller CH, Orians GH, Purves WK and Hillis DM. Life: The Science of Biology 8th Edition. Massacheusetts and Virginia: Sinauer Associates Inc. and W.H. Freeman and Company, 2008. Print.
Shafer RW, Stevenson D and Chan B. 1999. Human immunodeficiency virus reverse transcriptase and protease sequence database. Nucleic Acids Research 27: 348-352.
Upadhyay A, Pandey N, Mishra CA, Talele TT and Pandey VN. 2010. A single deletion at position 134, 135, or 136 in the beta 7-beta8 loop of the p51 subunit of HIV-1 RT disrupts the formation of a heterodimeric enzyme. Journal of Cellular Biochemistry 109: 598-605.
Works Cited (Information about thumb, fingers, palm, connection and RNase domains):
Kohlstaedt LA, Wang J, Friedman JM, Rice PA and Steitz TA. 1992. Crystal structure at 3.5 Angstrom resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science 256: 1783-1790. http://www.ncbi.nlm.nih.gov/pubmed/1377403
Kumar A, Kim HR, Sobol RW, Becerra SP, Lee BJ, Hatfield DL, Suhandolnik RJ and Wilson SH. 1993. Mapping of nucleic acid binding in proteolytic domains of HIV-1 reverse transcriptase. Biochemistry 32: 7466-7474. http://www.ncbi.nlm.nih.gov/pubmed/7687875
Yap S, Sheen C, Fahey J, Zanin M, Tyssen D, Lima VD, Wynhoven B, Kuiper M, Sluis-Cremer, N, Harrigan PR and Tachedjian G. 2007. N3481 in the connection domain of HIV-1 reverse transcriptase confers zidovudine and nevirapine resistance. PLOS Medicine 4: 1887-1990. http://www.ncbi.nlm.nih.gov/pubmed/18052601
Please direct questions or comments to Pallavi Penumetcha