One-Time Flippable DNA Segments for Biological Computers
by Bruce Henschen
Results
I compared the Hin-flipping activity of a HixM-HixC mutant construct with the Hin-flipping activity of a wt HixC-HixC construct.
1. FACS Data showed that pLac is a "leaky" promoter
It's variability makes it difficult to determine its orientation using phenotypes
2. Used DNA-Based Method of determining pLac flipping:
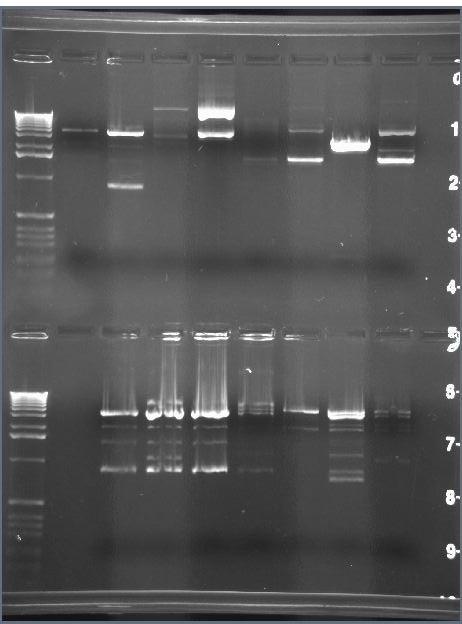
After flipping, cut the construct with PvuII:
If pLac points right, then cut out portion will be 886 bp (and cells should glow green):
If pLac points left, then cut out portion will be 780 bp (and cells should not glow green):
These results show that green cells (on left) correspond to cells that have 886 bp fragments (on right), but they also show no bands in the expected range for Hix Mutant colonies.
 |
Conclusions:
1. pLac is not frozen in its final conformation after Hin is selected against.
2. Hin displays altered activity on plasmids with one-bp insertions in the left Hix site - possible DNA damage?
Further Study:
Develop another method to determine whether Hix sites enabled Hin to flip DNA.
Perform flipping analysis on additional Hix mutants.
Pair Hix mutants with other Hix mutants and observe Hin activity.
Biology Main Page
© Copyright 2007 Department of Biology, Davidson College, Davidson, NC 28036