![]()

The computer program MacDNAsis was used to analayze the sequences of phosphofructokinse (PFK) that I obtained for the last assignment (see my Genbank web page). The results of that analysis are summarized here, through a number of figures.
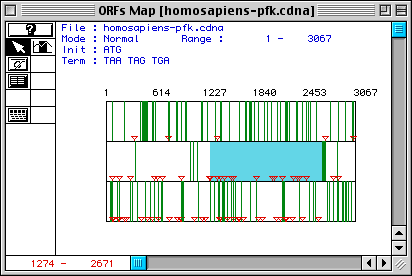
Figure 1. Largest open reading frame in Homo sapiens PFK cDNA.
The three rows correspond to the three possible reading frames of the sequence.
Red triangles represent start codons, and the green bars demarkate stop codons.
The red numbers at the bottom-left corner of the image indicate that the longest
ORF in Homo sapiens PFK cDNA sequence
is from nucleotides 1274-2671. To see DNA sequences of PFK, you can link to
my Genbank web page or click on the figure. By using
MacDNAsis to trasnlate this ORF to an amino acid sequence, I found the predicted
molecular weight of phosphofruktokinase to be 51,062.18 Daltons. According to
a printed source, this protein exists as a tetramer weighing approximately 340
kilodaltons (Stryer, 1996). Apparently there is some
discrepancy between the number I obtained using MacDNAsis and the protein's
weight in the literature.
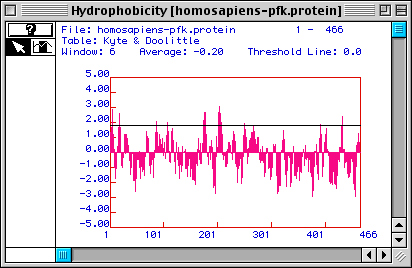
Figure 2. Kyte & Doolittle hydrophobicity plot of Homo sapiens
PFK. The translated ORF sequence (from Fig. 1) yielded this Kyte & Doolittle
hydrophobicity plot. Areas above the x-axis indicate hydrophobic portions of
the protein, and areas under the line represent hydrophilic ones. Parts of the
protein with values greater than 1.8 (approximated by the black line in this
figure) are good candidates for membrane-spanning domains. According to this
prediction, there appear to be at least 5 possible membrane-spanning regions.
However, judging from the function of PFK in the cell (an enzyme in the glycolytic
pathway), PFK should be a cytosolic protein (because glycolysis takes place
in the cytoplasm). The hydrophobic portions of the protein might be explained
as inner parts of the protein not exposed to the cytosol when PFK is in its
native conformation.
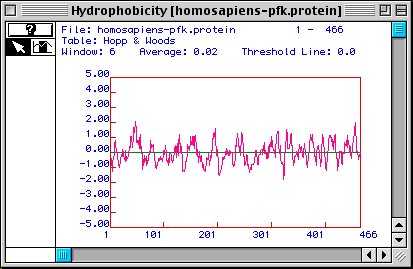
Figure 3. Hopp & Woods hydrophobicity plot of Homo sapiens PFK.
The Hopp & Woods plot differs from Kyte & Doolittle plots in that the
peaks in this figure correspond to hydrophilic portions of an amino acid sequence.
The highest peaks represent the most hydrophilic parts, which also tend to be
antigenic because they are likely to be on the outer surface of the protein's
native conformation. If one wanted to make a monoclonal antibody to a linear
epitope of the protein's tertiary structure, the most hydrophilic portion of
the sequence would be used. For this particular protein, the sequence from approximately
amino acids 33 to 66 seems to be the most ideal epitope, although many other
hydrophilic regions exist.
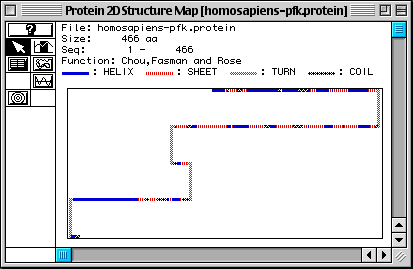
Figure 4. Chou, Fasman, and Rose predicted secondary structure of Homo
sapiens PFK. MacDNAsis predicted that numerous alpha helices (blue)
and beta pleated sheets (red) are present in PFK. There also seem to be a number
of coils (black), and four turns (green). This seems to be a good prediction,
because the actual 3D structure has many helices, sheets, and coils. To see
for yourself, look at a 3D Rasmol image of PFK by going to my Genbank
web page.
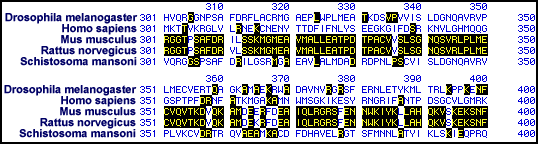
Figure 5. Multiple sequence alignment of various PFK sequences. A sample
of the alignment of PFK sequences from Drosphila
melanogaster, Homo sapiens, Mus
musculus, Rattus norvegicus,
and Schistosoma mansoni (click on the organism
name to see the original sequences obtained in my genbank
search). Residues highlighted in black indicate matches between sequences;
unhighlighted residues are not conserved between the species. The Mus
musculus and Rattus norvegicus
sequences seem to be very highly related. The sequences from other species show
a few similarities, but do not appear to be very related to the two aforementioned
sequences.
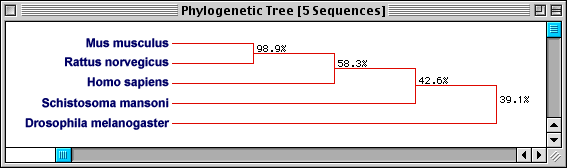
Figure 6. Phylogenetic tree of various PFK sequences. The phylogenetic
tree serves to predict the evolutionary relatedness of the species (Mus
musculus, Rattus norvegicus,
Homo sapiens, Schistosoma
mansoni, and Drosphila melanogaster)
on the basis of the percentage of amino acids conserved between their respective
PFK sequences. As predicted from Fig. 5, Mus musculus
and Rattus norvegicus are extremely
(98.9%) related, which is not surprising considering both organisms belong to
the subfamily Murinae. The Homo sapiens
sequence is the next most closely related species, which also agrees with a
taxonomical assessment (all these organisms are in the class Mammalia). The
two most distantly related species taxonomically, Schistosoma
mansoni and Drosophila melanogaster,
are also the least related in terms of sequence.
Davidson College Molecular Biology Homepage
Questions, comments - contact: jopalma@davidson.edu