This web page was produced as an assignment for an undergraduate course at Davidson College.
The largest open reading frame in human triosephosphate isomerase
(TIM) appears between animo acids 263-1117 (Fig.1).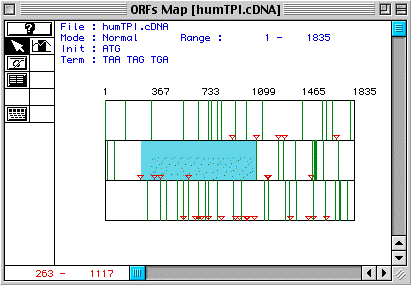
Figure 1. The largest ORF in the cDNA for human TIM occurs between amino acids 263-1117 (blue).
Follow this link to see the DNA sequence for human triosephosphate isomerase.
Using MacDNAsis, the predicted molecular weight for the human triosephosphate isomerase protein (based on the longest ORF of the cDNA) is 30,534.05 kDa. Interestingly, the German Heart Institute Berlin says that the molecular weight of human TIM is 26,538 kDa. This shows that the MacDNAsis program is not completely capable of predicting protein molecular weight based on DNA sequence information.
Based on the hydorpathy plot for human TIM (Fig. 2) generated by MacDNAsis, it is possible that human TIM is an integral membrane protein becuase there are four peaks that rise above 1.80 (which is the usual threshold for determining if a peak might or might not be an integral protein).
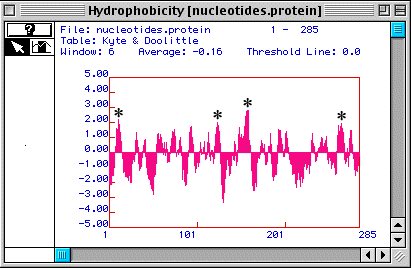
Figure 2. A Kyte & Doolittle Hydopathy Plot suggests that human TIM may be an integral membrane protein. Four peaks rise above 1.80 (*), suggesting four possible transmembrane domains.
Figure 3 shows a Hopp & Woods antigenicity plot for human TIM. This plot shows that the nucleotide domain marked with an asterisk is the best part of the protein to use to generate a peptide to make a monoclonal antibody against the peptide so that the antibody will recognize a linear epitope while allowing the protein to maintain its native conformation.
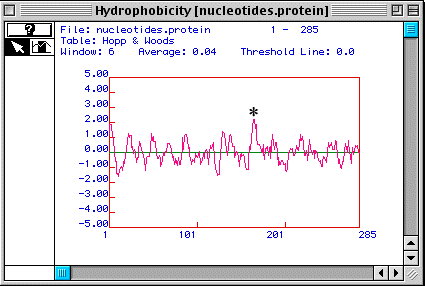
Figure 3. A Hopp & Woods antigenicity plot suggests a portion of the protein (*) that could be used to generate a peptide for use in making a monoclonal antibody to the peptide.
In Fig. 4, secondary structure is predicted for human TIM (Fig. 4), showing 16 alpha-helices and 18 beta-pleated sheets. However, we know that TIM is a protein comprised of eight alpha-helices surrounding eight beta-pleated sheets (see the CHIME image, a TIM dimer), in a barrel-like formation (Stryer, 1995). This shows that MacDNAsis is not always correct in its predictions about secondary structure.
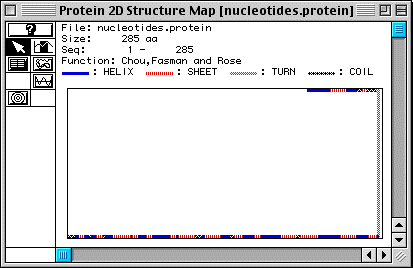
Figure 4. Predicted secondary structure of TIM .
MacDNAsis was also used to preform a multiple sequence alignment of TIM from five genome organisms (Fig. 4). This image shows sequence similarities between these organisms. Note that no spaces (-) have been added, so sequences may be match more closely than they appear.
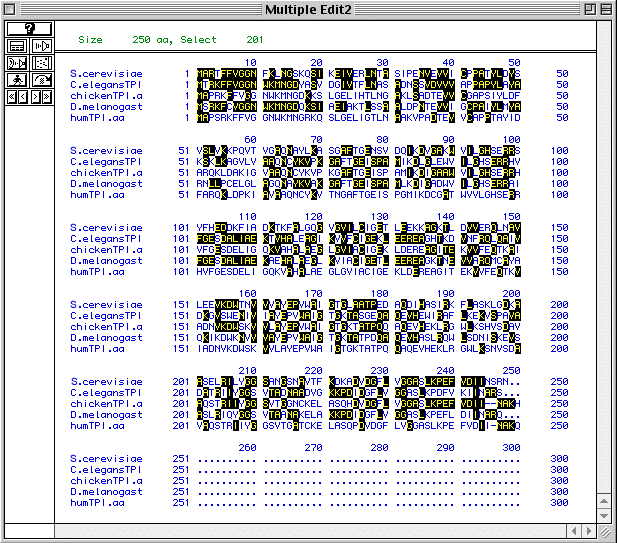
Figure 4. Multiple sequence alignment for five genome orgnaisms (S. cerevisiae, C. elegans, G. gallus, D. melanogaster, and H. sapiens). To see the individual protein sequences of TIM in these organisms, follow these links:
Figure 5 shows a phylogenetic tree of these five organisms based on the degree of amino acid conservation over time.
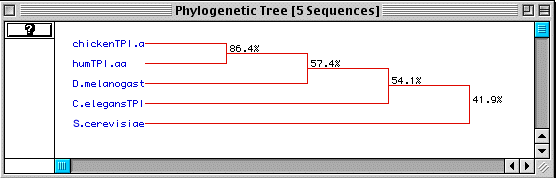
Figure 5. Phylogenetic tree of five genome organisms based on conservation of amino acid sequence over time.
Stryer, L. Biochemistry, fourth ed. W.H. Freeman and Co., New York: 1995. p 488.
Return to Luke's Molecular Biology Homepage
Return to Molecular Biology at Davidson College
© Copyright 2000 Department of Biology, Davidson College, Davidson, NC 28036
Send comments, questions, and suggestions to:luroberts@davidson.edu