Results
of MacDNAsis analysis of hexokinase
MacDNAsis is a computer program which can analyze DNA, RNA or
amino acid sequences to give a variety of information. The following information
was collected based on the yeast mRNA sequence for
hexokinase, which was found from Genbank.
1.
Determination of the largest ORF
To start the analysis of hexokinase, the mRNA sequence
of this protein in yeast was used to determine the ORF (open reading frame)
that encoded the protein.
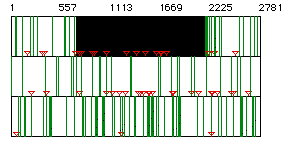 Figure 1: ORF for yeast hexokinase mRNA. Red triangles
indicate start codons, green lines indicate stop codons. The black
box represents the largest ORF for the mRNA sequence. Each row represents
a different reading frame and the resulting start and stop codons.
The ORF covers nucleotides 718 to 2178.
Figure 1: ORF for yeast hexokinase mRNA. Red triangles
indicate start codons, green lines indicate stop codons. The black
box represents the largest ORF for the mRNA sequence. Each row represents
a different reading frame and the resulting start and stop codons.
The ORF covers nucleotides 718 to 2178.
2.
Sequence Information
The ORF of yeast hexokinase mRNA was translated by MacDNAsis
to give the amino acid sequence. The program calculated the molecular
weight to be 53,949.57 kD. This sequence was used to predict
secondary structure of hexokinase and to prepare hydropathy and antigenicity
plots.
3.
Structural Information
The amino acid sequence was used to predict structural
information about hexokinase. Three types of analysis were performed
for hexokinase: a Kyte and Doolittle hydropathy plot, a Hopp and
Woods antigenicity plot, and a Chou, Fasman and Rose secondary structure
prediction.
-
Kyte and Doolittle hydropathy plots predict whether
the protein is a integral membrane protein. Negative numbers represent
hydrophilic regions; positive numbers represent hydrophobic regions.
A value greater than +1.8 suggests where transmembrane regions may lie.
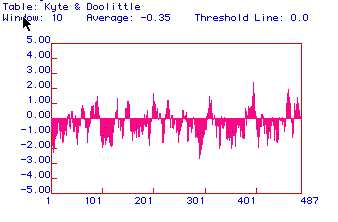 Figure 2: Kyte and Doolittle hydropathy plot for hexokinase.
This plot suggests there is one segment that crosses a membrane near amino
acid 400. Most other regions of the protein appear to be hydrophilic.
Figure 2: Kyte and Doolittle hydropathy plot for hexokinase.
This plot suggests there is one segment that crosses a membrane near amino
acid 400. Most other regions of the protein appear to be hydrophilic.
-
The Hopp and Woods antigenicity plot predicts areas
that may make good epitope sites. These sites are characterized by
greater hydrophilicity. Since hydrophilic regions are usually found
on the surface of the protein, the amino acids are better exposed to potential
antibodies. Hydrophilic regions are denoted by positive numbers;
hydrophobic by negative numbers.
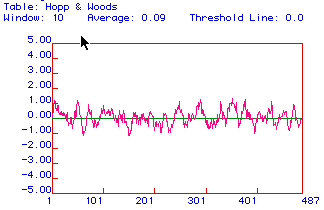 Figure 3: Hopp and Woods antigenicity plot for hexokinase.
No major hydrophilic regions are evident; however, the overall structure
appears to be more hydrophilic then hydrophobic. This prediction
agrees with that of the Kyte and Doolittle plot.
Figure 3: Hopp and Woods antigenicity plot for hexokinase.
No major hydrophilic regions are evident; however, the overall structure
appears to be more hydrophilic then hydrophobic. This prediction
agrees with that of the Kyte and Doolittle plot.
-
The Chou, Fasman and Rose function in MacDNAsis predicts
the secondary structure of proteins in a simplified linear form.
The analysis predicts a-helix,
b-pleated sheets,
turns and coils.
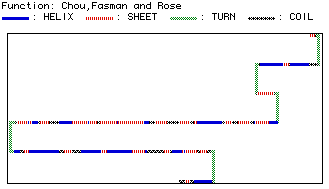 Figure 4: Predicted secondary structure of yeast hexokinase.
The tertiary
structure can be viewed with RasMol.
Figure 4: Predicted secondary structure of yeast hexokinase.
The tertiary
structure can be viewed with RasMol.
4.
Comparison among species
MacDNAsis was used to compare the hexokinase amino acid
sequence translated from yeast mRNA to the amino acid sequence found for
four other species. Genbank searches for Schistosoma
mansoni, Arabidopsis
thaliana, mouse
and human
provided amino acid sequences for hexokinase in each species. (To
view the results of that search, click on the corresponding species.)
-
Multiple sequence alignment was performed to determine the
genetic conservation among the five species. A sample of the alignment
is shown below.
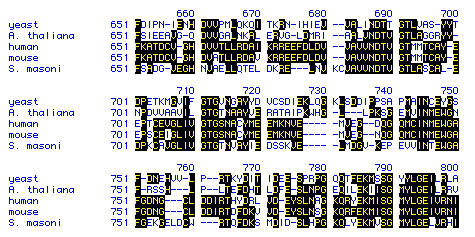 Figure 5: Segment of amino acid sequence of hexokinase for
each of the five species analyzed. Blackened letters indicate amino
acids that match the yeast sequence.
Figure 5: Segment of amino acid sequence of hexokinase for
each of the five species analyzed. Blackened letters indicate amino
acids that match the yeast sequence.
-
A phylogenic tree calculating the degree of homology between
species was generated after the sequence allignment.
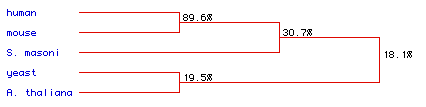 Figure 6: Phylogenetic tree for the five species in this analysis.
The degree of amino acid conservation between species is indicated by percentage
values. Strong conservation is seen between the human and mouse sequences
(89.6%).
Figure 6: Phylogenetic tree for the five species in this analysis.
The degree of amino acid conservation between species is indicated by percentage
values. Strong conservation is seen between the human and mouse sequences
(89.6%).
return
to Genbank Search page
return to my home page
© Copyright 1999,
Department of Biology, Davidson College, Davidson, NC 28036
Send comments, questions,
and suggestions to: lafreeman@davidson.edu






