Affinity Chromatography is a relatively simple, yet quite effective molecular technique which isolates antibodies, antigens, hormones, or other proteins by taking advantage of their binding affinity for their respective ligands. In the following explanation, I shall first describe the materials required for a successful isolation, then walk through the actual procedure, before finally discussing briefly the chromatogram.
Materials (after explanation in TAMUK web site)
The major materials required for an affinity chromatography procedure are 1) a bead matrix, 2) a ligand, 3) a solution containing the substrate to be isolated, 4) a wash to elute the non-bound impurities in the solution, and 5) a final wash to elute the bound substrate from its ligand.
The bead matrix is an agarose gel loaded into an elution column. Sepharose is the most widely used matrix, because the hydroxyl groups on the sugar residues can be easily manipulated to accept a ligand.
The ligand is then selected according to the desired isolate. For example, if you wanted to isolate antibodies specific for antigen A from an antiserum, you would choose antigen A as your ligand. Likewise, if you wanted to isolate a specific enzyme, you could use either its substrate, an inhibitor, or even a cofactor. Next, one simply needs a mixture containing the desired isolate. Two factors are required for the ligand: First, that it bind specifically and reversibly to the isolate. Ideally, the ligand should have an affinity for its substrate between the range of 10-4 and 10-8M in free solution. Secondly, that the ligand is capable of covalent bonding to the matrix without disrupting its binding activity. This is usually facilitated by the placement of spacer arms between the ligand and the matrix, so that in case the active site is buried deep within the ligand, it is not physically hidden from it's binding substrate.
The solution is usually a protein rich mixture such as antiserum, which is poured into the elution column and allowed to run through the gel, at a controlled rate.
The first wash must be of sufficient salt concentration, pH, or temperature to elutes all unbound impurities from the solution, but not so extreme that it causes the isolate to dissociate from the ligand. The second wash however, must elute the isolate, therefore, it must be sufficiently extreme in concentration, pH, or temperature.
Procedure (after Janeway and Travers 1997, and Stryer 1995)
In short, affinity chromatography relies on isolating the desired protein from a mixed solution through the protein's specific binding affinity to ligands mounted in a gel matrix through which the protein mixture allowed to run. In the following paragraphs, I shall break this procedure into four steps, which are summarized in Fig. 1.

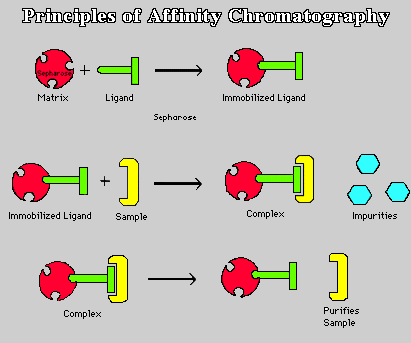
Fig. 1. Cartoon illustration of steps for affinity chromatography. Top panel: bonding of ligand to Sepharose matrix results in immobilized ligand-matrix. Middle panel: Sample mixture is added to gel column. Complimentary proteins bind to ligand-matrix complex, while the non-binding impurities are washed out. Bottom panel: In order to remove the bound proteins and retrieve them from the gel, a stronger second wash is run through. Your favorite protein is then washed out. (Figure taken from <http://ntri.tamuk.edu/fplc/pursammat.html>.)
1) Binding of the selected ligand to the matrix requires that a covalent bond be formed between the two (Fig. 1, top panel). As mentioned above, this is facilitated by derivitization of the sugar residues' hydroxyl groups. Also as mentioned above, it is important to realize that the substrate might not be able to reach the ligand active site if it is hidden deep within the ligand. Therefore, most ligands are attached first to spacer arms which are then bonded to the matrix. The ligand-matrix gel is then loaded into an elution column.
2) Once the column has been prepared, the mixture containing your favorite isolate is poured into the elution column (Fig. 1, middle panel). Once in the column, gravity pulls the solution through the gel, because most of the proteins do not bind to the ligand-matrix complex. However, when the ligand's recognized substrate passes through the gel, it binds to the ligand-matrix complex, halting its passage through the gel. Some of the impurities flow through the gel due to gravity, but most remain, unbound, in the gel column.
3) In order to remove these unbound impurities, a wash of extreme pH, salt concentration, or temperature is run through the gel (Fig. 1, middle panel). It is important to use a strong wash so that all the impurities are removed, but it is also just as crucial that the wash be not so strong that it removes the bound isolates. Once the impurities are washed-out, the only remaining part of the protein mixture should be the desired isolates.
4) Finally to collect your favorite isolate, which is still bound to the ligand-matrix in the gel, a stronger second wash is run through the column (Fig. 1, bottom panel). This second wash relies on the reversible binding properties of the ligand, which allows the bound protein to dissociate from its ligand in the presence of this stronger wash. The protein is then free to run through the gel and be collected.
You have now isolated your favorite protein!
The Chromatogram (after Carrier and Bordonaro 1994)
Before finishing, I ought to briefly mention the chromatogram. Affinity chromatography is not just limited to isolating one protein; given a sample of similar proteins with similar binding affinities, a chromatogram can be generated. A chromatogram is a plot of absorbance vs. time for the elution of proteins in affinity chromatography. Carrier and Bordonaro write that four things may be learned from a chromatogram: 1) the level of complexity of the sample (indicated by the number of peaks) 2) qualitative information about the sample composition (by comparing peak positions with known standards) 3) quantitative information of the relative component concentrations (by comparing peak areas) and 4) total column performance (by comparing with known standards).
Figure 2 may help in illustrating some of this information. It shows four different elutions, for four different pH washes, each from a different column. Each sample mixture contained four proteins. As the pH increases, it is clear that the elution of drops in intensity and also increases in time of elution so that for a wash of pH 9.0, protein 1 has overlapped signals with protein 2. The elution of proteins 3 and 4 vary in intensity over the different pH washes with no apparent trend, although they elute out at around 3 and 4 minutes, respectively. Although not exhaustive, this is a sample of the type of information which can be read from a chromatogram.
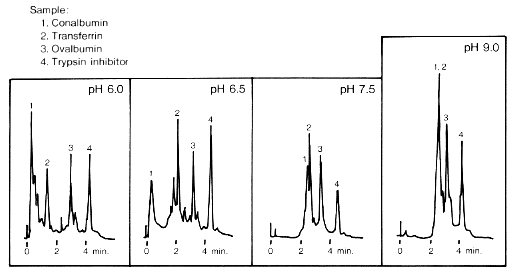
Fig 2. Four chromatograms from four separate elutions at varying levels of pH (6.0, 6.5, 7.5, and 9.0). Each sample mixture contained the four proteins (preceded by corresponding numbers): 1. conalbumin; 2. tranferrin; 3. ovalbumin; and 4. trypsin inhibitor. The pH washes were the following: pH 6.0 25mM Piperazine-HCl; pH 6.5 25mM Piperazine-HCl; pH 7.5 25mM Tris-HCl; and pH 9.0 25mM Ethanolamine-HCl. Flow rate was 1.5 mL/min. Absorption readings were measured at 280 nm. (Figure taken from <http://www.sdk.co.jp/shodex/english/dc010503.htm>.)
References
Carrier R and Bordonaro J. 1994. Chromatography-The Chromatogram. <http://www.rpi.edu/dept/chem-eng/Biotech-Environ/CHROMO/firstpg.html>
Janeway CA and Travers P. 1997. Immunobiology. 3rd ed. London: Current Biology Limited/Garland Publishing Inc. p 2:10-2:11.
Stryer L. 1995. Biochemistry. 4th ed. New York: W.H. Freeman and Company. p 50.
TAMUK (Texas A&M University-Kingsville). Affinity Chromatography. <http://ntri.tamuk.edu/fplc/affin.html> Accessed 1998 Feb. 14.
TAMUK (Texas A&M University-Kingsville). Principle of Affinity Chromatography. Affinity Chromatography. <http://ntri.tamuk.edu/fplc/pursammat.html> Accessed 1998 Feb. 14.
![]()
Please send your comments, suggestions to grnoland@davidson.edu.
Back to GSN's homepage.
Link to Davidson College's Biology Homepage.