A very popular reporter gene and protein has been chloramphenicol acetyltransferase or CAT. This is a bacterial gene that evolved to neutralize the antibiotic chloramphenicol. To neutralize it, CAT transfers acetyle groups to chloramphenicol and thus changes its shape into a harmless form. Molecular biologists were quick to recognize this enzyme could be used as a reporter protein in eukaryotic cells. If you wanted to determine when a promoter were activated, you could place the promoter of interest upstream of the CAT coding sequence and either measure mRNA or protein production. Eric Davidson measured both mRNA and protein production in the chapter on single gene circuitry.
Figure 1. Chime interactive view of CAT enzyme structure. b-mercaptoethanol is shown in the chloramphenicol binding site. The active form of CAT is a homotrimer, but only the monomer is shown here.
Measurement of mRNA was performed by in situ hybridization. To perform the enzyme assay, you must first extract the proteins from the cells and then mix this extraction with chloramphenicol that has been synthesized with radioactive hydrogens (3H). In addition, the substrate acetyl CoA is also added to the mixture. The amount of acetylation is directly proportional to the amount of CAT enzyme present. Therefore, you can measure the amount of acetylated chloramphenicol in different protein extractions and determine how much protein was produced as a result of activated promoters. CAT can add two acetyl groups to each chloramphenicol.
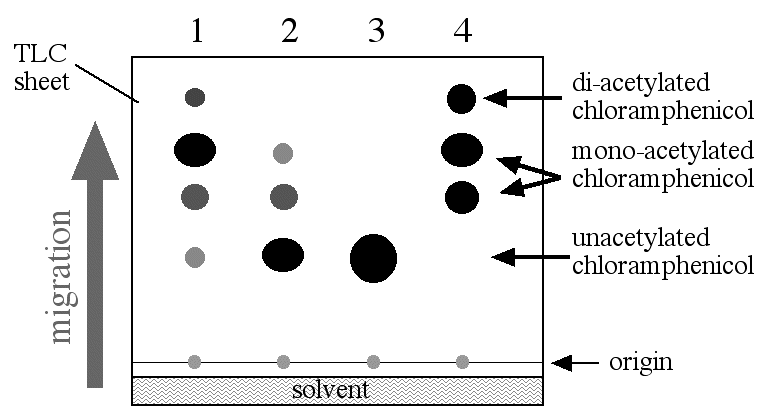
When the CAT reactions is completed, the products are placed on thin-layer chromatographic (TLC) sheets, placed in the appropriate solvent (a mixture of chloroform and methanol) and allowed to migrate up the TLC sheet which is exposed to X-ray film (figure 2). Each product (two forms of chloramphenicol with one acetyl group and one with two acetyl groups added) and all the unused substrate (acetyl CoA, chloramphenicol and CAT) will migrate on the TLC surface according to their ability to interact with surface of the TLC. Since the only radioactive molecule is chloramphenicol, the only molecule visible on the X-ray film is chloramphenicol and all its acetylated forms.

Figure 2. CAT assay results. The reaction mixture was placed on a TLC sheet and lowered into the TLC chamber with a little bit of solvent in the bottom. The reaction mixture was spotted onto the origin and allowed to migrate up the TLC sheet. When the solven reached the top of the TLC sheet, it was stopped and allowed to dry. Later, it was exposed to X-ray film which was developed. The spots shown in this figure would only be visible on the X-ray film and the level of the solvent would not be visible. Figure 2 was a composit to show all the parts in a signle diagram. Lane 1: promoter 1 construct; promoter 2 construct; negative control; postive control.
The amount of acetylated chloramphenical can be measured either by scanning the density of the dark spots on the X-ray film or by measureing the radioactivity in each spot on the TLC sheet. Either way, CAT assays offer an indirect but quantitative way to measure the amount of transcription driven from any given promoter. In figure 1 you can see two differnent strength promoters were tested in addition to the two controls. The amount of acetylated chloramphenicol is greater in lane 1 than lane two and therefore promoter number 1 was able to produce more mRNA than promoter number 2.
© Copyright 2002 Department of Biology, Davidson College, Davidson, NC 28036
Send comments, questions, and suggestions to: macampbell@davidson.edu