This page will summarize the results of our efforts to clone all 5 yeast IDH-encoding genes.
First Experiment (Tuesday Lab; 5 Feb., 2002):
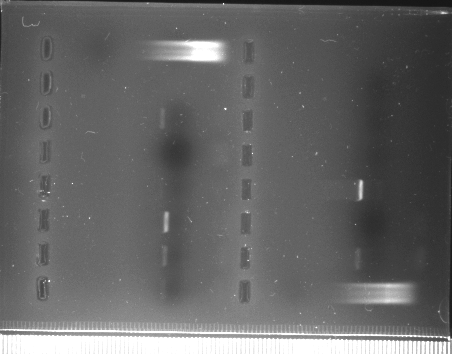
Below are two gels show the reults of three MgCl2 concentrations for each of the five genes. For each gene, 1.5, 1.75 adn 2.0 mM MgCl2 was used in the reactions with 50 µL the final volumes and hotstart was used in all cases by heating the block to 95° C before placing the tubes in the thermocycler. These three concentrations were loaded next to each other from left to right when the wells are rotated to the top. You can download the PDF file for the DNA molecular weight markers called "1 kb ladder" produced by Gibco/BRL which is now a part of Invitrogen.

Top Row: IDH1, IDP3, blank, degraded MW markers.
Bottom Row: degraded MW markers, IDP2, IDH2.
0.7% agarose gel, 0.5X TBE.

IDP2, blank, degraded MW markers, Fresh MW markers
loaded later.
0.7% agarose gel, 0.5X TBE.
First Experiment (Thursday Lab; 7 Feb., 2002):
We attempted to build on Tuesday's results by scaling up the volumes. However, I mistakenly increased the number listed for yeast genomes per microgram of DNA and so the lab diluted their template 100X too much. In addition, the volumes were all increased to 100 µL from Tuesday's 50 µL reactions.
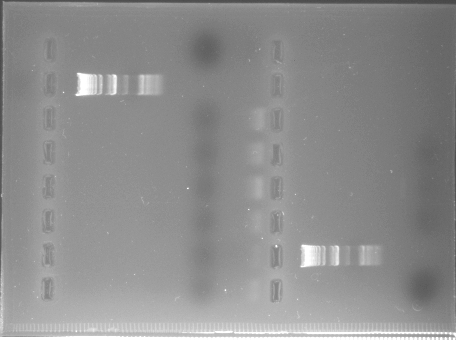
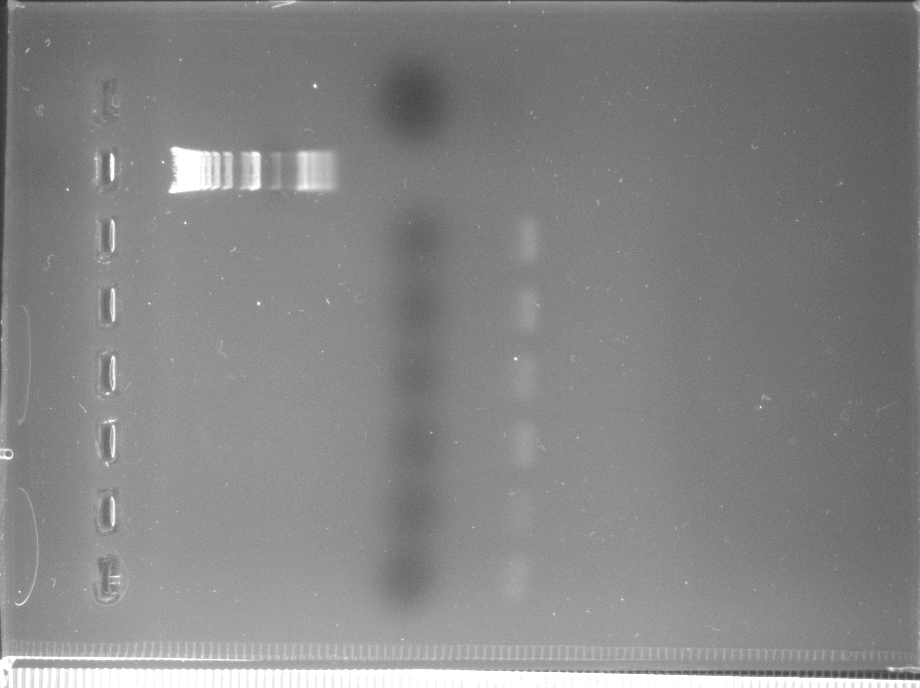
All PCR lanes showed no amplification occurred.
Trouble Shooting Experiment (Friday Volunteers; 8 Feb., 2002):
In class, it was decided that the volume change may have played a role since the anneling temperature was 40° C.
In addition, I remembered that the two lab sections had very different dilutions of templates so I contacted a few members of Tuesday's lab and they told me they either used the template undiluted (IDH1, IDP1, and IDP2) or diluted 10X (IDP3). Therefore, template from Nona and Tricia was used directly for all subsequent reactions.
The gel below shows the results when reaction volumes were 100 µL (top row) or 50 µL (bottom row). MgCl2 was held at 2.0 mM and hotstart was used again.
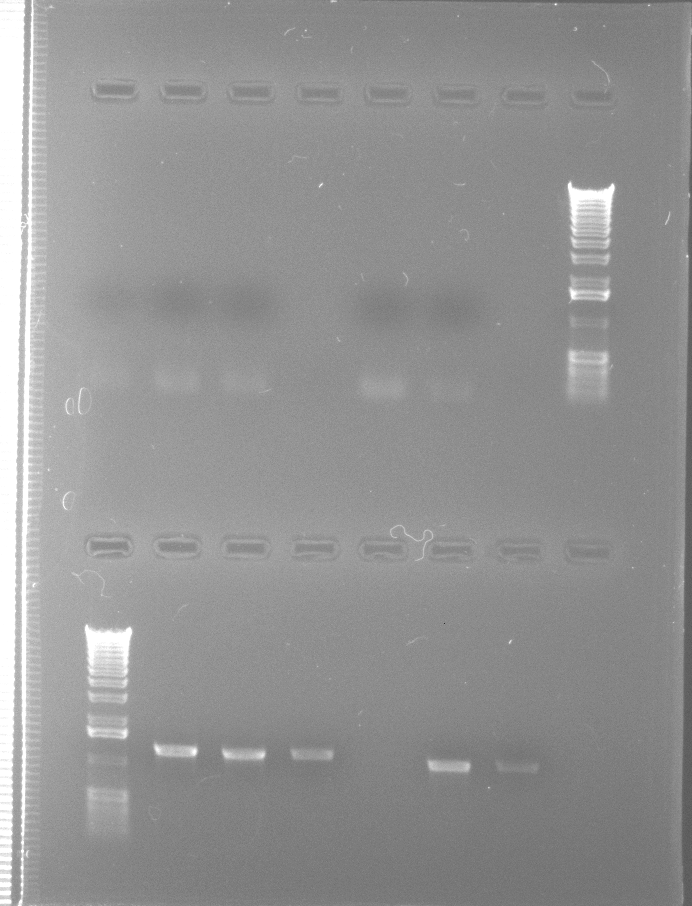
TopRowof 100 µL reactions: IDP1,
IDP2, IDP3, blank, IDH1, IDH2, blank, 1 kb ladder.
Bottom Row of 50 µL reactions: 1 kb ladder, IDP1, IDP2, IDP3, blank,
IDH1, IDH2.
0.7% agarose gel, 0.5X TBE.
Scale Up Experiment (Saturday 9 Feb., 2002):
To insure each group would have enough PCR product for cloning, more reactions were performed using 2.0 mM MgCl2 and hotstart with 50 µL volumes. Each gene was done in duplicate, pooled, and 8 µL was used to load on the gel below. In addition, the IDH2 gene was amplified using 2.25 mM MgCl2 to see if it would work better at a slightly higher MgCl2 concentration.
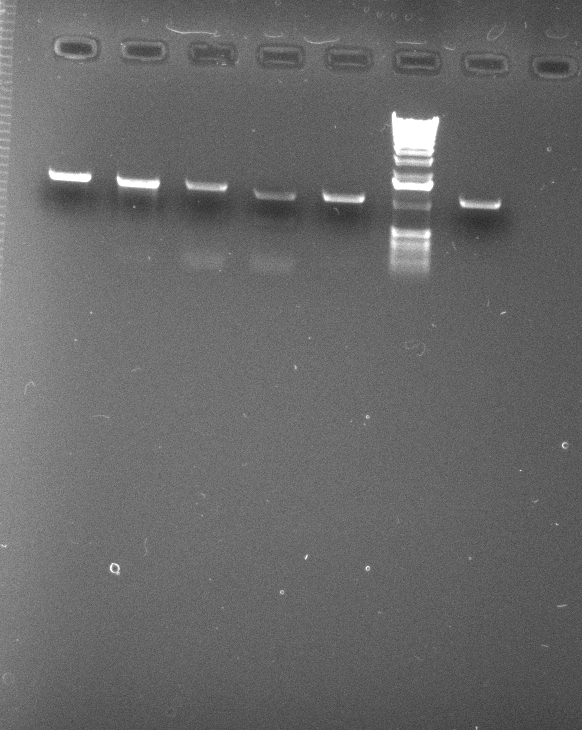
50 µL reactions of: IDP1, IDP2, IDP3, IDH1, IDH2, 1 kb ladder, IDH2 with 2.25 mM 2.0 MgCl2.
0.9% agarose gel, 0.5X TBE.
Each group will have about 45 µL of PCR product to purify for the cloning step.
Lab Schedule In Context of Research Project