Proc. Natl. Acad. Sci. USA
Vol. 84, pp. 9123-9127, December 1987
Genetics
( position effects / P transposon / enhancers / ß-galactosidase / cell-type markers )
Dquartment of Cell Biology, Biozentrum, University of Basel, Klingelbergstrasse 70, CH-4056 Basel, Switzerland
Communicated by Walter J. Gehring, September 4, 1987
ABSTRACT We have developed an approach for the in situ detection of genomic elements that regulate transcription in Drosophila melanogaster. The approach is analogous to a powerful method of bacterial genetics, the random generation of operon fusions, that enables the isolation and characterization of genes simply by knowing or postulating their pattern of expression; it is not necessary initially to screen for mutant phenotypes. To apply this approach to Drosophila, we have used the expression of the lacZ gene of Escherichia coli from the P-element promoter in germ-line transformant flies to screen for chromosomal elements that can act at a distance to stimulate expression from this apparently weak promoter. Of 49 transformed lines obtained, ~70% show some type of spatially regulated expression of the lacZ gene in embryos; many of these express lacZ specifically in the nervous system. The P-lacZ fusion gene is, therefore, an efficient tool for the recovery of elements that may regulate gene expression in Drosophila and for the generation of a wide variety of cell-type-specific markers.
The random generation of operon fusions in prokaryotes involves integrating a promoterless reporter gene, whose expression can easily be detected and assayed at many deferent positions in a target genome, so that it comes under the control of a random selection of chromosomal promoters (e.g., ref. 1 and 2). This approach enables the isolation and characterization of genes, simply by knowing or postulating their pattern of expression, it is not necessary initially to screen for mutant phenotypes. In eukaryotes, however, for the efficient expression of the reporter gene, not only would an active promoter be required, but the reporter gene would normally have to be the first gene in the fusion transcript. We, therefore, decided to consider a different approach to the development of an analogous tool in eukaryotes.
Position effects, i.e., dependence of expression levels on the genomic position of an integrated gene, have frequently been reported in eukaryotes (e.g., ref. 3). This sensitivity of promoter activity to adjacent genomic sequences suggested to us that a suitable detection system would allow us to detect genomic elements that could regulate transcription at a distance. We, therefore, chose to express a reporter gene (in this case the lacZ gene of Escherichia coli, which codes for ß-galactosidase) from a weak promoter and to integrate this promoter fusion by P-element transformation into the Drosophila genome. Thus, an increase in expression of the reporter gene should reflect the activity of nearby enhancer-like elements in the genome that can act at a distance on the weak promoter.
In this work we report the results obtained using a fusion of lacZ to the P-element promoter. By staining embryos from fly lines transformed with the P-lacZ fusion gene for ß-galactosidase activity, we can visualize many cell-type- and tissue-specific patterns of lacZ expression. The P-lacZ fusion is, therefore, an efficient tool for the detection of elements that regulate gene expression in Drosophila and for the generation of a wide variety of cell-type markers. The range of molecular and genetic approaches that can be applied to Drosophila facilitates the subsequent isolation and characterization of the fly genes that may also be associated with the regulatory elements recovered.
MATERIALS AND METHODS
DNA Manipulations. DNA manipulations were performed essentially as described (4). Restriction enzymes, DNA polymerase I, and T4 DNA ligase were obtained from Biofinex (Praroman, Switzerland). Calf intestinal alkaline phosphatase and DNase I were obtained from Boehringer Mannheim. For cloning steps, a high yield of linearized partially EcoRI-digested vector was obtained by digestion in the presence of ethidium bromide (5) followed by agarose gel isolation of the linear digestion product. in situ hybridization of the biotinylated plasmid pLacA92 to polytene chromosomes was performed as described (6). 5-Biotin (19)-2 ' deoxyuridine 5'-triphosphate was obtained from Calbiochem (Luzern, Switzerland). Detek I-hrp (Enzo Biochemicals, New York), a complex of streptavidin and biotinylated horseradish peroxidase, was used for detection of hybridized probe.
Germ-Line Transformation. ry506 (ORM)
embryos (3) 0-90 min. of age were injected as described (7) with a DNA solution
containing pLacA92 (300 µg/ml) and p25.7wc (100 µg/ml)
(8).
ß-Galactosidase Localization. ß-Galactosidase localization in embryos was performed as described (3) except that the staining reaction was carried out at pH 7.0 at 37°C. 5-Bromo-4-chloro-3-indolyl ß-D-galactoside was obtained from Bachem Fine Chemicals (Bubendorf, Switzerland). Dimethylformamide was removed from the aqueous staining solution before use by placing on ice for 5-10 min and then centrifuging for 1 min. in an Eppendorf centrifuge to remove the precipitate.
Drosophila Strains. ry506(ORM) flies were obtained from Y. Hiromi (Stanford University). The following balancer stocks, obtained from Y. Hiromi, were used to establish and maintain transformant lines: FM6; ry506 CyO; ry506 and TM3, ry, Sb. For a full description of marker genes and balancer chromosomes see Lindsley and Grell (9).
RESULTS AND DISCUSSION
We hypothesized that the promoter used to express lacZ on the P-element should have three properties. First, it should be relatively weak, so that an increase in its activity in some or all cells can easily be detected. Second, it should be active constitutively in all cells throughout development, so that any spatially regulated pattern of expression due to nearby enhancers is superimposed on a low uniform background. Third, it should lie at one end of the P-element, so that it can be relatively exposed to the influence of enhancers on at least one side of the integration site.
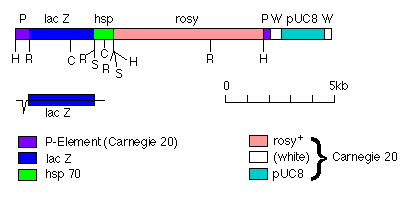
FIG. 1. Plasmid pLacA92 carrying the transposon P[lac,ry+]A. The plasmid is depicted as linear, with the 5' end of the P element at the left. A 0.9-kilobase (kb) EcoRI fragment carrying the 3' end of the lacZ gene and the stop codon and polyadenylation site of the Drosophila hsp70 gene (3) was cloned into the EcoRI site at position 590 of Carnegie 20 (10) giving pLac33 (data not shown). A 3.0-kb EcoRI fragment carrying the rest of the lacZ gene was then cloned into the EcoRI site at position 590 of pLac33. The orientations and positions of the two cloned EcoRI fragments were confirmed by using the restriction sites shown. The end of the lacZ fragment on the 5' side of the central Sac I site is derived originally from pSKS106 (11), and its insertion as an EcoRI fragment into the polylinker of Carnegie 20 should result in an in-frame translational fusion to the second exon of the P element. The end of the lacZ gene on the 3' side of the Sac I site is derived originally from pUR288 (12). The origins of the other parts of pLacA92 are also shown. Underneath the restriction map the deduced structure of the fusion transcript is shown. Restriction endonuclease sites: H, Hind III; R, EcoR I; S, Sal I; C, Sac I. There are other Sac I sites in the rosy gene whose positions have not been mapped.
A promoter that appeared to fulfill these criteria was the P-element promoter itself, whose main transcription start site is 87 base pairs from the 5' end of the element (8). We, therefore, constructed the transposon P[lac,ry+]A (Fig. 1), which contains (i) an in-frame translational fusion of lacZ to the second exon of the P element, (ii) both ends of the P element that are necessary in cis for transposition, (iii) the rosy (ry ) gene as a marker, and (iv) on the 3' side of lacZ, the trailer sequences and the polyadenylation site of the Drosophila hsp70 gene.
The plasmid carrying the transposon, pLacA92, was injected into ry506 (ORM) Drosophila embryos (3) along with a helper
P element, p25.7wc (8); the latter produces transposase but is itself
incapable of transposing. Of 171 injected survivors (the G0
generation), 39 gave rise to one or more ry+ transformants in the
next generation (G1); in all, 348, ry+
G1 flies were obtained. All transformant offspring
(between 1 and 30) of each G0 survivor were
pooled and intercrossed (or backcrossed to ry- flies if transformants
were of one sex only) to obtain second generation (G2)
flies. Embryos were collected overnight from the 39 G2pools,
fixed, and stained for ß-galactosidase activity using 5-Bromo-4-chloro-3-indolyl
ß-D-galactoside.
FIG. 2. Embryonic ß-galactosidase
staining patterns of five different P[lac,ry+]A insertion strains.
Embryos were fixed, stained, mounted whole, and photographed using Nomarksi
phase-interference microscopy. b is a dorsal view; j is a ventral view.
In all other photographs dorsal is on the top, and ventral is on the bottom.
Anterior is on the left in all photographs. Embryos are ~500 µm long.
In all five cases the presence of a single insertion was confirmed by in
situ hybridization to larval salivary gland chromosomes. All five insertions
are homozygous viable with no obvious phenotype. Abbreviations: AP, anal
pads; B. brain; C, cephalic sensory organs; CL, clypeolabrum; CNS central
nervous system; CPS, cephalopharyngeal skeleton; HG, hindgut; Ich5, lateral
abdominal pentascolopidial chordotonal organ; MG, midgut; P. proctodeum;
PMG, posterior midgut; PS, posterior spiracle; PV, proventriculus; S, stomodeum;
Y, yolk; Md, Mx, Lb, T1, mandibular, maxillary, labial, and first thoracic
segments.
(a and b) Endogenous ß-galactosidase activity of ry506
embryos at stages 15 and 17 respectively, as defined by Campos-Ortega and
Hartenstein (13). Expression begins at stage 14 in a line of cells along
the dorsal midline. These cells later appear to disperse throughout much
of the embryo.
(c and d) Embryos carrying insertion 46 (cytological map position 42A/B),
at stages 5 and 10 of embryogenesis, respectively. ß-Galactosidase
expression begins (arrowheads) while cellularization of the blastoderm is
proceeding and is detectable at 10% to 90% of egg length. In later embryos
there is little or no detectable expression in the stomodeum and posterior
midgut, tissues that are derived from the cells that do not stain at blastoderm
(14).
(e and f) Embryos carrying insertion 49 (X chromosome-linked) at stages
11 and 17, respectively. ß-Galactosidase expression begins in vitellophages
at stage 9 and in the developing central nervous system at stage 11. In
later embryos the vitellophages are incorporated with the yolk into the
midgut, and weaker staining also appears in the region of the cephalic sensory
organs and in a number of cells posteriorly that we have not been able to
identify.
(g and h) Embryos carrying insertion 5 (cytological map position 56F) at
stages 11 and 17, respectively. ß-Galactosidase expression begins
at stage 10 in segmentally repeated ventral ectodermal stripes. The stripes
show a two-segment periodicity in the strength of expression and are in
the posterior part of each segment. Other sites of expression at this stage
are in the proctodeum, clypeolabrum, and the procephalic neurogenic region
(not in focus). In later embryos the segmentally repeated pattern occurs
in the central nervous system and appears to be continued into the brain.
Other sites of expression include the cephalic sensory organs, the anal
pads, a group of cells at the base of each posterior spiracle, cells surrounding
the cephalopharyngeal skeleton, and a single cell laterally in the epidermis
of each abdominal segment (not in focus); this may be either the lateral
td or lateral bd neuron of Bodmer and Jan (15).
(i) Detail of two adjacent abdominal segments from a stage-17 embryo carrying
insertion 18 (cytological map position 85A). The insertion stains many cells
in the central nervous system (data not shown) and the peripheral nervous
system, including at least three cells in each unit of the lateral pentascolopidial
chordotonal organ (13, 16). (Bar = 50 µm)
(j) Embryo carrying insertion 30 (cytological map position 61F) at stage
17. ß-Galactosidase expression begins at stage 10 in the stomodeum,
proctodeum, and segmentally repeated epidermal stripes. At stage 11 the
cells bounding the labial lobe begin to express. In later embryos expression
is strongest in the proventriculus, the anal pads, posterior hindgut, and
some of the cephalic epidermis. Weaker sites of expression include epidermal
cells adjacent to segment boundaries, the heart, midgut, and remaining hindgut.
Approximately 20% (8/39) of the pools gave rise only to embryos with no detectable ß-galactosidase activity except for the endogenous pattern of staining (Fig. 2 a and b). Levels of E. coli ß-galactosidase staining in these embryos were apparently below the limit of detection with 5-bromo-4-chloro-3-indolyl ß-D-galactoside under the conditions used, and these pools were not analyzed further. The other 80% (31/39) of G2 pools gave rise to mixtures of lac+ and lac- embryos. The lac+ embryos had many different spatial patterns of lacZ expression (Fig. 2), and some pools obviously contained several patterns. The lac+ embryos were expected because of the presence of ry-flies and ry+ heterozygotes in the G2 pools.
Several individual G2flies from each of the 31 pools were used to breed stocks homozygous for each P[lac,ry+]A insertion (or balanced heterozygous stocks if the chromosome carrying the insertion was homozygous lethal). In all, 49 different insertion strains with different patterns of lacZ expression were bred from these individuals. Evidently, multiple insertions had occurred in many injected G0 embryos. Of the 49 lines, 12 expressed lacZ in all or most of the embryo, and 37 clearly showed some sort of spatially regulated expression of lacZ.
Some examples of the patterns recovered are shown in Fig. 2. Twenty-three strains expressed lacZ specifically in either the central or peripheral nervous system. Seven of these strains expressed lacZ in all or most of the central nervous system (e.g., Fig. 2 e and f); 13 showed expression in distinctive subsets of cells in the central nervous system (e.g., Fig. 2 g and h); and at least 8 showed expression in the thoracic and abdominal peripheral nervous system (e.g., Fig. 2i). These will be described in more detail elsewhere. Thirteen strains expressed lacZ specifically in some part of the gut. These included strains that expressed the enzyme specifically in the pharynx, proventriculus, midgut, or hindgut (e.g., Fig. 2). Other sites of lacZ expression found included the epidermis (Fig. 2 c, d, and j), malpighian tubules, muscles, and amnioserosa (data not shown).
At least four strains expressed lacZ initially in a two segment periodicity (e.g., Fig. 2 g and h). The frequency of such insertions, and the fact that none of them mapped to locations of known pair-rule or segment-polarity mutations (refs. 17-19, and data not shown) suggest that most of these patterns are a consequence of the segmentation process and not due to cis-acting control elements of neighboring segmentation genes. In this model most of these patterns would be markers for early differentiation events within each segment that are controlled directly or indirectly by the segmentation genes.
Ten out of 49 insertions (20%) were homozygous lethal. Four of these insertions expressed lacZ in all or most of the embryo; 6 expressed it in a clear spatially regulated manner. Hence, in most cases, the P-lacZ fusion can come under the control of Drosophila expression elements without causing major disruption to the structure or expression of an adjacent essential gene. Other workers have reported a preference of P elements for genomic targets in promoter regions, and some such insertions appear to alter the level or pattern of expression of the downstream transcript without abolishing it (20, 21). Thus, many P insertions near promoters and in intergenic regions may bring the P-lacZ fusion under the control of at least some of the elements regulating expression of one or more adjacent Drosophila genes without giving a lethal phenotype.
We expect the expression elements that we detect to be enhancer-like elements that can act at a distance on the P promoter. It may be possible in some cases that there is transcriptional readthrough from Drosophila sequences into the P element and that the ATG initiation codon of the P-lacZ fusion gene is fortuitously the first ATG codon capable of initiating translation. In such cases the fusion gene would be expressed from a Drosophila promoter rather than from the P promoter, though it is difficult to envisage this situation occurring very frequently.
Our results suggest that expression of the P promoter is very dependent on sequences adjacent to the integration site. In light of this, it is not surprising that restriction of transposase expression to the germ line is regulated post-transcriptionally, at the level of RNA splicing (22, 23).
An important question to be asked is whether there are Drosophila genes adjacent to the P[lac,ry+]A insertions that are also regulated by the elements specifying the expression patterns of the P-lacZ fusion gene. In at least some cases we might expect to find a nearby transcript with a similar expression pattern to that of the P[lac,ry+]A insertion.
The dominant ry+ marker on the P[lac,ry+]A element also allows a genetic approach to finding adjacent genes that may be necessary for normal functioning of the cells that express lacZ. If insertion stocks are treated with a mutagen that generates deletions (e.g., x-rays), ry- revertants that may also have neighboring genes deleted can be recovered in one generation.
We have shown that the P[lac,ry+]A transposon is, at the very least, an efficient tool for the generation of specific cell markers. These markers should be useful for a variety of genetic and developmental studies on the marked cells. If the patterns recovered also reflect the expression patterns of adjacent Drosophila genes, P[lac,ry+]A should also be very useful for the recovery of genes that are important for the functioning of particular cell-types. In particular, it should facilitate dissection of the Drosophila nervous system at the morphological, genetic, and molecular levels. In principle, the system should also be applicable to other eukaryotic organisms.
We thank Yash Hiromi for performing most of the injections and Hugo Bellen for performing Southern blot hybridizations and critical reading of the manuscript. We also thank Clive Wilson for critical reading of the manuscript and Stephan Schneuwly for plasmids carrying the fragments that were subcloned. C.O'K. is the holder of a long-term fellowship from the European Molecular Biology Organization. This work was supported by the Swiss National Science Foundation and the Cantons of Basel.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" in accordance with 18 U.S.C. §1734 solely to indicate this fact.
References
1. Casadaban, M. J. & Cohen, S. N. (1979) Proc. Natl. Acad. Sci. USA 76, 4530-4533.
2. O'Kane, C., Stephens, M. A. & McConnell, D. (1986) J. Bacterial 168, 973-981.
3. Hiromi, Y., Kuroiwa, A. & Gehring, W. J. (1985) Cell 43, 603-613.
4. Maniatis, T., Fritsch, E. F. & Sambrook, J. (1982) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Laboratory, Cold Spring Harbor, NY).
5. Parker, R. C., Watson, R. M. & Vinograd, J. (1977) Proc. Natl. Acad. Sci. USA 74, 851-855.
6. Langer-Safer, P. R., Levine, M. & Ward, D. C. (1982) Proc. Natl. Acad. Sci. USA 79, 4381-4385.
7. Spradling, A. C. & Rubin, G. M. (1982) Science 218, 341-347.
8. Karess, R. E. & Rubin G. M. (1984) Cell 38, 135-146.
9. Lindsley, D. L. & Greil, E. H. (1968) Genetic Variations of Drosophila melanogaster (Carnegie Institute of Washington, Washington, DC).
10. Rubin, G. M. & Spradling, A. C. (1983) Nucleic Acids Res. 11, 6341-6351.
11. Shapira, S. K., Chou, J., Richaud, F. V. & Casadaban, M. J. (1983) Gene 25, 71-82.
12. Ruther, U. 8~ Muller-Hill, B. (1983) EMBO J. 2, 1791-1794.
13. Campos-Ortega, J. A. & Hartenstein, V. (1985) The Embryonic Development of Drosophila melanogaster (Springer, Berlin).
14. Technau, G. M. & Campos-Ortega, J. A. (1985) Wilhelm Roux's Arch. Dev. Biol. 194, 196-212.
15. Bodmer, R. & Jan, Y.-N. (1987) Wilhelm Roux's Arch. Dev. Biol. 196, 69-77.
16. Ghysen, A., Dambly-Chaudière, C., Aceves, E., Jan, L.-Y. & Jan, Y.-N. (1986) Wilhelm Roux's Arch. Dev. Biol. 195, 281-289.
17 Nüsslein-Volhard, C., Wieschaus, E. & Kluding, H. (1984) Wilhelm Roux's Arch. Dev. Biol. 193, 267-282.
18 Jürgens, G., Wieschaus, E., Nüsslein-Volhard, C. & Kluding, H. (1984) Wilhelm Roux's Arch. Dev. Biol. 193, 283-295.
19. Wieschaus, E., Nüsslein-Volhard, C. & Jurgens, G. (1984) Wilhelm Rowux's Arch. Dev. Biol. 193, 296-307.
20. Tsubota, S., Ashburner, M. & Schedl, P. (1985) Mol. Cell. Biol. 5, 2567-2574.
21. Kelley, M. R., Kidd, S., Berg, R. L. & Young, M. W. (1987) Mol. Cell. Biol. 7, 1545-1548.
22. Laski, F. A., Rio, D. C. & Rubin, G. M. (1986) Cell 44, 7-19.
23. Rio, D. C., Laski, F. A. & Rubin, G. M. (1986) Cell 44, 21-32.
Return to Molecular Biology Main Page
© Copyright
2000 Department of Biology, Davidson College, Davidson, NC 28036
Send comments, questions, and suggestions to: macampbell@davidson.edu