MacDNAsis of Myosin cDNA
MacDNAsis is a software program which allows cDNA and amino acid sequences to be analyzed. From this program, we can find the largest open reading frame in the cDNA, predict 2d shape and hydrophobicity of the encoded protein, and compare the protein encoded by the cDNA to the same protein in other species. These analyses were performed on the protein myosin, specifically on cDNA encoding myosin from mice.

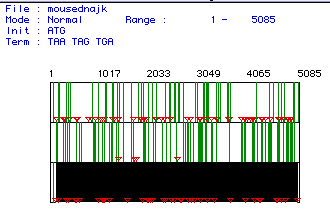
Figure 1. Open Reading Frame. This panel shows the open reading frames for the mouse cDNA encoding myosin. The three panels indicate three different reading frames. The red triangles indicate the start codon and the green bars indicate the stop codon. The longest open reading frame is highlighted in black and was the one used for this analysis. This open reading frame occurs between bases 105 and 5084.
To see the sequence for mouse cDNA and amino acids, click here.
From analysis, I found the molecular weight of myosin for mice to be 191.56 kiloDaltons. The MacDNAsis analyzed the amount of each amino acid present and determined the molecular weight (data not shown).
Figure 2. Two Dimensional Structure of Myosin. This figure predicts the two dimensional structure of the mouse protein myosin. Blue segments represent an alpha helical shape, red segments a beta pleated sheet, green segments a turn, and black segments a coil. From this analysis, it appears that myosin is composed primarily of amino acids arranged in a helical formation. We can compare this predicted structure with the RasMol image of myosin (To see the RasMol image, click here. ). We see in the RasMol image a long segment of alpha helix which has a turn in the middle. This segment is predicted in the 2d structure analysis by the long blue helix containing a green turn in the middle of the diagram. The RasMol image indicates a large percentage of alpha helix and turns, with fewer beta pleated sheets and coils. This is seen in the 2D prediction as well.
Figure 3. Kyte-Doolittle Hydrophobicity Plot. This figure is a Kyte-Doolittle hydrophobicity analysis of the mouse protein myosin. It is a prediction of structure. Regions of the protein below the 0.0 line are hydrophilic and regions above the line are hydrophobic. Peaks above two occur where there are transmembrane domains. Based upon this analysis, we can predict that myosin is predominantly hydrophilic and that there are no transmembrane domains in myosin. This is what we would expect as we know that myosin is not located in cellular membranes.
Figure 4. Hopp Woods Hydrophobicity Plot. This figure is a Hopp-Woods analysis of the mouse protein myosin. The Hopp-Woods table predicts hydrophilic regions of the protein. The highest peaks are the most hydrophilic and therefore likely to be more antigenic. This means that it is easier to make a monoclonal antibody specific to this region of the protein. Based on the plot, I would try to make an antibody to the region of the protein at either amino acid number 1100 or amino acid number 1400.
Figure 5. Amino Acid Sequence Homology. This is an illustration of amino acid sequence homology in myosin for the species chicken, Drosophila, C.Elegans, human, and mouse. Highlighted regions indicate sequence conservation. As illustrated in the figure, there are numerous regions where sequence similarity is conserved, as in amino acids 730 to 740. However, there are also regions where there is little to no sequence conservation, as in amino acids 810 to 820. This figure is not very useful in determining the percentage of homology between these sequences, however. For this reason, we will use a phylogenetic tree analysis.
Figure 6. Phylogenetic Tree. A phylogenetic tree showing degrees of similarities between myosin amino acid sequence in the five chosen species. From this figure, we see that chicken and human myosin share 87.3 percent of amino acids. Of these, 44.1 percent are found in Drosophila. Of the sequences shared between these three, 37.1 percent are also found in C.Elegans. Finally, of the amino acids conserved between these four species, 24.3 percent are also found in mice. From this plot, we can conclude a surprising fact; that human myosin is more homologous to chicken myosin than it is to that of another mammal, the mouse. However, upon closer examination, it appears that this sequence similarity is due in large part to different sizes of amino acid chains being analyzed. The human amino acid sequence is shorter than that of the mouse which decreases the amount of sequence similarity present. The next step would be to analyze a portion of the mouse amino acid sequence similar in size to that in the mouse and see if there is a higher percentage of sequence conservation.
To link to the cDNA and amino acid sequence for myosin in mice, click here.
To link to the amino acid sequence for myosin in humans, click here.
To link to the amino acid sequence for myosin in Drosophila, click here.
To link to the amino acid sequence for myosin in chicken, click here.
To link to the amino acid sequence for myosin in C. Elegans, click here.
To return to my homepage, click here.