Molecular Biology Homepage
by Susan Schultz
last updated 04/9/98
RasMol
Image of Taq Polymerase
MMDB Id: 5779 PDB Id: 1TAU
The Wonderful World of Antisense Technology
From DNA to Protein Expression: Protein molecules are the expressions of a gene. However, to get to a protein the cell must undergo two complex processes, transcription and translation. Transcription is the process in which an RNA copy is made of the DNA. In order to get the copy, many enzymes such as polymerase, helicase, exonuclease, ligase and single stranded binding proteins work together to unwind the double helix and match the base pairs of RNA (adenine, guanine, cytosine and uracil) to the DNA. Once the copy is made, the RNA molecule, which is now in the heterogeneous nuclear RNA (hnRNA) mode, is still not ready to go express the gene by making a protein. The hnRNA must be spliced to remove non coding sequences, and protected from the cellular environment with a 5' cap and poly- A tail. Finally, the hnRNA is transported out of the nuclear membrane and into the cytoplasm where it achieves the status of mRNA. In the cytoplasm, the mRNA molecule hooks up with ribosomes where the protein production can start. Every three nucleotides in the mRNA molecule codes for a specific amino acid and are appropriately called a codon. The codon pairs with an anti codon of tRNA that has attached to an amino acid. In this manner a polypeptide chain is formed. It will eventually twist and contort itself into a unique configuration which aids in the function of the protein. Occasionally, a bad mRNA molecule is synthesized so that the resulting protein can not function properly. Abnormalities of proteins cause many diseases that afflict humans. Therefore, it seems logical to conclude that if the expression of these malfunctional proteins could be stopped, the sources of disease would be obliterated and the disease will be treated, if not cured. This idea is the basis for antisense technology.
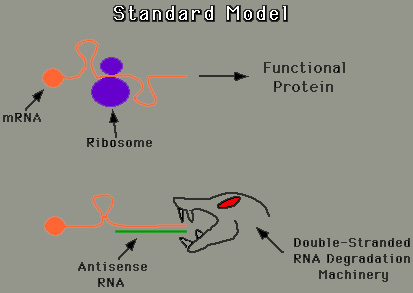
The Basics of Antisense: A sense strand is a 5' to 3' mRNA molecule or DNA molecule. The complementary strand, or mirror strand, to the sense is called an antisense (What is Antisense 1998). Antisense technology is the process in which the antisense strand hydrogen bonds with the targeted sense strand. When an antisense strand binds to a mRNA sense strand, a cell will recognize the double helix as foreign to the cell and proceed to degrade the faulty mRNA molecule, thus preventing the production of undesired protein. Although DNA is already a double stranded molecule, antisense technology can be applied to it, building a triplex formation (Fritz 1996).
A DNA antisense molecule must be approximately seventeen bases in order to function, and approximately thirteen bases for an RNA molecule. RNA antisense strands can be either catalytic, or non catalytic. The catalytic antisense strands, also called ribozymes, which will cleave the RNA molecule at specific sequences. A non catalytic RNA antisense strand blocks further RNA processing, i.e. modifying the mRNA strand or transcription (Fritz 1996).
The exact mechanism of an antisense strand has not been determined. The current hypotheses include blocking RNA splicing, accelerating degradation of the RNA molecule, preventing introns from being spliced out of the hnRNA, impeding the exportation of mRNA into the cytoplasm, hindering translation, and the triplex formation in DNA (Fritz 1996). The two figures below illustrate the fact that no consenus has been reached concerning how antisense accomplishes the reduction of protein synthesis.

Figure by CMello Labs
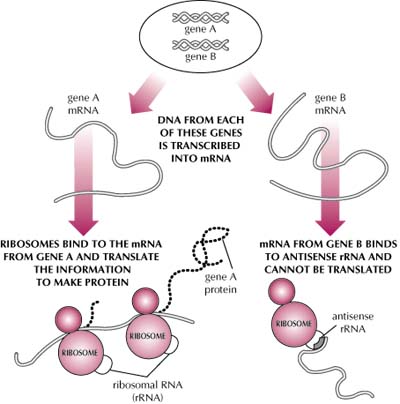
Figure by Teri Platt, courtesy of Fred Hutchinson Cancer Research Center
Inserting Antisense into Cells:
Endocytosis- One of the simplest methods
to get nucleotides in the cell, it relies on the cell's natural process
of receptor mediated endocytosis. The drawbacks to this method are the
long amount of time for any accumulation to occur, the unreliable results,
and the inefficiency (Dantus 1997).
Micro- Injection- As the name implies, the
antisense molecule would be injected into the cell. The yield of this method
is very high, but because of the precision needed to inject a very small
cell with smaller molecules only about 100 cells can be injected per day
(Dantus 1997).
Liposome Encapsulation- This is the most effective
method, but also a very expensive one (Dantus 1997). Liposome encapsulation
can be achieved by using products such as LipofectACE(TM)
to create a cationic phospholipid bilayer that will surround the
nucleotide sequence. The resulting liposome can merge with the cell membrane
allowing the antisense to enter the cell.
Electroporation- The conventional method of adding a nucleotide sequence to a cell can also be used. The antisense molecule should traverse the cell membrane after a shock is applied to the cells.
Obstacles of Antisense: The
successful result of any of these methods is reduced protein production
as long as the surrounding conditions are favorable (Driver). However,
there are more problems that need to be overcome then just inserting the
antisense molecule into the cell. If antisense is being used as a treatment
in the human body, it might be degraded before stopping any protein production
if it is unmethylated because the body would recognize it as an invader
(Dantus 1997). When antisense is used in living organisms, there are also
many complications that can arise such as high blood pressure and a low
white blood cell count (Dantus 1997).
Applications of Antisense: Applications
of antisense technology are very diverse. It was first successfully used
in plants. One example of antisense technology that is currently available
is the Flavr Savr tomato. Antisense was used to block the enzyme that is
involved in spoilage, thereby increasing the length of time a tomato could
be sold.
Antisense technology is now used in mammalian cells. Promising fields of
study for antisense technology in humans include cancer gene therapy and
AIDS. In cancer treatment, antisense is constructed in a way that it will
bind with the mRNA from the PKC alpha gene (Conrad 1997). This gene is
targeted because PKC alpha kinase is more sensitive in cancer cells. The
treatment has resulted in a 50% decrease in the size of the tumor from
ovarian cancer in one patient and stabilization of the tumor's growth in
other patients (Conrad 1997). Tests are still being administered to determine
the possibility of future use of the antisense treatment on a wider scale
since the previous trial has shown antisense to be well- tolerated (Conrad
1997). See below for connections to other sites for more information.
Works Cited
Conrad, Eugene. June 1997. "Antisense Thearpy (AT) Blocks
Cancer Growth." <http://www.meds.com/conrad/asco/sikic.html>
Accessed 1998 Feb 18.
Dantus, Marcos. Fall 1997. "Antisense Olgionucleotide Technology in
AIDS Research." <http://www.msu.edu/~mackertm/introns/>
Accessed 1998 Feb 17.
Driver, Sam. "Answers to Frequently Asked Questions About Antisense."
<http://www.ummed.edu/pub/c/cmello/AFAQ.html>
Accessed 1998 Feb 3.
Fritz, Jeff. 07/29/96. "Antisense Technology." <http://www.mc.vanderbilt.edu/gcrc/gene/antisens.htm>
Accessed 1998 Feb 3.
O'Leary, Michael. 1996. "AntisenseDiagram." <http://www.fhcrc.org/about/CenterNews/1996/Jul3/Antisensediagram.htm>
Accessed 1998 Feb 3.
"What is Antisense?" 02/10/98. <http://www.hybridon.com/graphic_version/antisense/stepbystep.html>
Accessed 1998 Feb 3.
If you would like to know more about the use of antisense as a treatment,
please click on you area of interest:
Cancer Treatment
Fred
Hutchinson Cancer Research Center
HIV/ AIDS
If you would like to know more about Davidson's biology classes:
Molecular Biology Home Page
© Copyright 2000 Department of Biology, Davidson College,
Davidson, NC 28036
Send comments, questions, and suggestions to: suschultz@davidson.edu