This webpage was produced as an assignment for an undergraduate course at Davidson College.
Protein Kinase C Orthologs
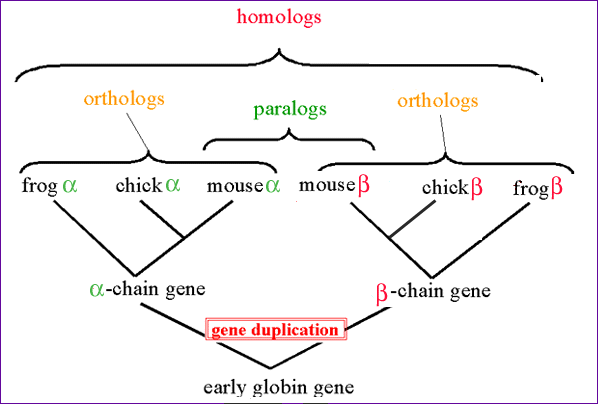
Figure 1: Image depicts concepts of Orthology and paralogy. Obtained from NCBI. Permission Pending.
Introduction
Evolution is the theory that all organisms or forms of life have adapted from a single common ancestor. This development is best categorized by the small variance of change seen between species that can be traced back, connecting different groups through their shared characteristics - noting the subtle changes that elicit tremendous differences. One way that biologists can trace this change is through studying gene orthologs. An ortholog is simply a gene that shares a common ancestor (Campbell, 2005). By analyzing conserved regions in a gene, valuable information can be learned about a protein and the evolution of that protein and/or gene throughout time. This webpage aims at identifying various orhtologs of protein kinase C and by comparing their variations, obtaining a better understanding of how PKC has developed over time.
Eukaryotes:
Protein kinase C (PKC), as noted on the structure and function page, comprises of a family of kinase proteins that help mediate signal transduction cascades by hydrolyzing lipids. As shown before, this family of kinases varies in sequence between lysozymes and thusly plays different roles relative to their unique affinities. While PKC varies in complexity in eukaryotes, from the single isoform found in S. cerevisiae (budding yeast) to the 12 found in mammals, all isoforms share a highly conserved carboxyl terminal domain (Rosse et al., 2010). This region then serves as a marker for evolutionary adaptation because its presence can be traced between both orthologs and paralogs that developed due to speciation; leaving the regulatory domain of the gene as the principal factor in variation. In order to better understand the development of PKC through gene orthology, let us focus specifically on Human PKCα rather than the entire superfamily of PKC.
Human (Homo sapiens) PKCα is one of three types of conventional PKCs activated by binding of DAG and Ca2+ to its C1 and C2 conserved domains (Rosse et al., 2010). This particular kinase however, is known to play roles in processes such as cell adhesion, cell transformation, and volume control (Protein-Protein Kinase C). By using a BLAST to compare conserved domains found in the catalytic region of PKCα, Figure 2 was created showing the conserved and varying sequences of several orthologs of Human PKCα. This data proved to be highly conserved as supported by the literature. Uniquely though the regions encoding the activation loop, designated by the box below has conserved ends but unique central amino acid sequence. This difference in coding could account for the variance in protein function among the different orthologs caused by an alteration in shape of the activation site. This phenotypical difference could allow for the binding of a variety of ligands or even affect the overall function of the protein caused by activation. Also, because of PKCα's conserved ATP binding sites located at the beginning of Figure 2, AAs 345, 346, 350, etc. (Conserved Domains), new screens are being created that focus on these sites because of their utilization as kinase-specific inhibitors used in disease treatments (Li et al., 2004).
Conserved Catalytic Domains
Figure 2: Above figure shows the conserved regions for the catalytic domain (C3 & C4)of PKCalpha amino acids of various orthologs in comparison with Human PKCalpha. The Regulatory domain was ignored for this comparison because of large variance due to ligand specificity between species. For a full comparison of entire PKCalpha sequence click here. Red letters illustrates conserved sequences while Blue illustrates variances from the Human PKCalpha sequence. Boxed sequence is the location of the activation loop (A-loop). Sequences obtained from NCBI. Permission Pending.
While the catalytic region maintains a very important role in cellular functioning, the binding sites located upstream in the regulatory region play the most important role in regulating protein activation as well as function. Referring back to Figure 2 of the structures and functions page, the C1 and C2 coding regions serve as the primary binding sites of ligands Ca2+, DAG and PS. These ligands are pivotal for PKCα activation. Because of which, variances in these regions could account for different affinities of these ligands, affecting the overall output of PKCα activation. Knowing that the C2 region of PKCα allosterically interacts with Ca2+, inferences can be made as to the evolution of this sequence based upon Ca2+ interaction in skeletal systems (B. van Rossum and Patterson, 2009). Knowing that both Caenorhabditis and Drosophila are invertebrates, their differences in amino acid sequences in the C2 region suggests the less frequent use of Ca2+ mediated activation due to the lack of skeletal systems in these species (Figure 3).
ClustalW2 Comparison of PKCα Orthologs
Figure 3: Above figure compares PKCα amino acid sequences of Homo sapiens (humans), Mus musculus (house mouse), Gallus gallus (chicken), Caenorhabditis elegans (round worm), and Drosophila virilis (fruit fly) using a ClustalW2 comparison thanks to EBI. Conserved regions are designated by a * below the sequence, while a . and : designate variances in either one or two orthologs from that of human PKCα respectively. Regulatory domains C1 & C2 lie within their respective markers, the Activation Loop sequence lies within the boxed area, and the  designates the turn motif phosphorlyation site. According to the sequence, Mus musculus is extremely closely related to the Homo sapien gene and is thusly commonly used as a model organism in which valuable information can be obtained. Sequence data was acquired by performing a BLAST of the Homo sapiens PKCα protein FASTA sequence (NP_002728.1), courtesy of NCBI. Permission Pending.
designates the turn motif phosphorlyation site. According to the sequence, Mus musculus is extremely closely related to the Homo sapien gene and is thusly commonly used as a model organism in which valuable information can be obtained. Sequence data was acquired by performing a BLAST of the Homo sapiens PKCα protein FASTA sequence (NP_002728.1), courtesy of NCBI. Permission Pending.
Conclusion:
Protein kinase C α and its family of conventional PKCs are found in numerous species other than those presented here. However when searching for PKCα presence in E. coli, S. cerevisiae, and A. thaliana, similar proteins that served relatively parallel functions were found instead of PKC itself (other than S. cerevisiae which maintains a different form of PKC not discussed here - link to other orthologs). This led to the discovery that some less complex organisms contained in fact a particular lysozome of PKC, instead of a multitude as in the case of mammalians (Rosse et al., 2010). For this reason assigning a specific function to PKC presents difficulties because of the multitude of possible variances between sequences of orthologs and paralogs. However through genetic experiments performed on highly conserved species such as the "house mouse" a better understanding of PKCs subtle roles is becoming increasingly possible. Such experiments have led to findings such as those by Gwark et al. revealing that stimulation of PKCα suppresses colon caner formation through the down regulation of beta-catenin (2009). Other diseases have also been studied using PKC and its family of Ser/Thr kinases. A short list of these diseases and their correlating protein can be found at SMART.
While evolutionarily PKC has developed from the conserved catalytic region shared by kinase enzymes, to the multitude of subfamilies now comprising this superfamily of proteins, substantial differences can be seen by tracing the conservation of gene orthology between various species. Although specific functions have yet to be identified for all isoforms of PKC, increasing evidence supports nonredundant roles for many members of this unique family.
Works Cited
B. van Rossum D. and Patterson R. 2009. PCK and PLA2: Probing the complexities of the calcium network. Cell Calcium 45: 535-545. Science Direct.
BLAST. Basic Local Alignment Search Tool. http://blast.ncbi.nlm.nih.gov/Blast.cgi.
ClustalW2. EBI. http://www.ebi.ac.uk/Tools/clustalw2/index.html.
Gwark J. et al. 2009. Stimulation of protein kinase C-alpha suppresses colon cancer cell proliferation by down-regulation of beta-catenin. Journal of Cellular and Molecular Medicine 13: 2171-80. Pubmed.
Li B. et al. 2004. Creating Chemical Diversity To Target Protein Kinases. Combinatorial Chemisty & High Throughput Screening 7: 453-72. Pubmed.
Protein-Protein Kinase C. NCBI. http://www.ncbi.nlm.nih.gov/.
Rosse C. et al. 2010. PKC And The Control Of Localized Signal Dynamics. Nature Reviews Molecular Cell Biology 11: 103-112. NatureReviews.
Schultz et al. 1998. Letunic et al. 2008. S_Tkc. SMART .
William's Homepage
PKC Structure/Function
Molecular Biology Homepage
Please send any questions or comments to wigreen@davidson.edu