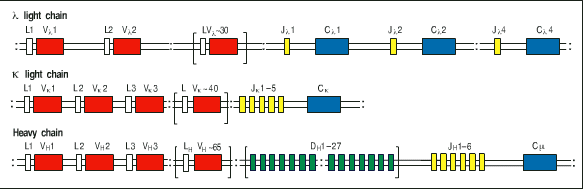
Figure taken from ImmunoBiology by Janeway et al., Forth Edition.
There is no time limit on this test, though I have tried to design one that you should be able to complete within 3 hours, except for typing. You are not allowed to use your notes, or any books, any electronic sources, nor are you allowed to discuss the test with anyone until all exams are turned in at 9:30 am on Monday February, 14. EXAMS ARE DUE AT CLASS TIME ON MONDAY FEBRUARY 14. You may use a calculator and/or ruler. The answers to the questions must be typed on a separate sheet of paper unless the question specifically says to write the answer in the space provided. If you do not write your answers on the appropriate pages, I may not find them unless you have indicated where the answers are.
-3 pts if you do not follow this direction.
Please do not write or type your name on any page other than this
cover page. Staple all your pages (INCLUDING THE TEST PAGES)
together when finished with the exam.
Name (please print here):
Write out the full pledge and sign:
Here is the honor code
http://www.davidson.edu/student/redbook/honorgeneral.html#honorcode
"On my honor I have neither given nor received unauthorized information regarding this work, I have followed and will continue to observe all regulations regarding it, and I am unaware of any violation of the Honor Code by others."
How long did this exam take you to complete (excluding typing)?
I. Define these terms - 2 pts each. When the term is preceded by an asterisk (*), provide a specific example to further demonstrate your knowledge. These terms can be define succinctly so using a lot of words is not the best way to demonstrate your fluency with these terms.
1) b barrel
- two beta sheets on
one protein connected by disulphide bonds, cylindrical in shape.
2) MIIC- endosomal compartment, MHC II is found here and loaded
with peptide as CLIP is produced and then removed.
3) * MHC restriction- TCR binds to peptide which is recognized only in the
context of sitting in an MHC binding groove. Example = TCR cannot
recognize the same peptide sitting in a different MHC allotype,
even in the same person.
4) * sandwhich ELISA- a method
where Ab is bound to well, antigen is added and binds, second
Ab binds and is detected via enzymatic reaction to produce colored
product. Example = quantification of the level of cytokine secretion
by some cells.
5) RAG-1- half of the RAG1/RAG2
complex used during somatic recombination, cuts at the 3' end
of the 7mer of the RSS, cuts only one strand of dsDNA.
6) CLIP- Ii chain is cut twice
in MIIC to produce CLIP after MHCII a
and b have assembled in ER and move on to
MIIC. CLIP is located in the binding groove of MHCII and acts
as chaperone to keep MHCII from unfolding until antigenic peptide
is loaded.
7) immunohistochemistry- method for labeling proteins in tissue
with antibody that is attached to an enzyme and produces a colored
product which is visible when viewed through a microscope.
8) P-nucleotides- Variation
produced during somatic recombination when the phosphodiester
backbone of nucleotides on one strand near the hairpin loop is
cut so that the nucleotides on this same strand are flipped over
the hairpin "hinge". The exact number of P nucleotides
produced is random and become a part of the coding joint.
9) somatic hypermutation- point
mutations producted after B cells have produced a functional Ig
and the B cells have been activated. The mutations occur randdomly
but are concentrated in the hypervariable regions and change the
affinity either higher or lower.
10) HLA-DM- MHCII-like instructure,
this molecule stablelizes the a/b MHC II in the MIIC as CLIP is removed and peptide
loaded by HLA-DM.
11) * hapten- small molecule
two which you want to make an antibody. Hapten is bound to a larger
carrier protein to stimulate an immune response since hapten is
too small to. Example = cAMP bound to lysozyme.
12) * recombination signal sequence-
RSS is used for somatic recombination and is comprised of a sequence
of nucleotides in this pattern: 7mer - spacer of 12 or 23 - 9mer.
Example = D segment - RSS (12)-intron - RSS(23) - J segment for
H chain somatic recombination.
13) MHC haplotype- the specific
allelic combination located in the MHC complex (includes about
200 genetic loci) on one chromosome
14) proteasome- always found
in the cytoplasm of a cell, this structure is a protease that
digests proteins in the cytoplasm. In lymphocytes, it can be modified
by LMP's to produce peptide fragments that are optimized for loading
in MHC I.
15) invariant chain- Chaperone that that helps in the assembly and folding
of MHC II a/b
in the ER. Ii is found as homotrimer and binds 3 MHC II heterodimers
to forma a megacomplex as a 9mer. Once the 9mer is assembled,
it leaves the ER and moves onto the MIIC. Ii also functions to
prevent premature peptide binding in MHC II.
More thoughtful questions:
6 pts.
1) How many MHC molecules with different specificity does
a human B cell express? Explain your answer.
Over 14.
3 MHC I genes and two chromosomes = 6 different MHC I on B cell.
4 MHC II genes (due to one a and two
b in HLA-DR); two chromosomes = 8 different
MHC II on B cell.
Plus, each of the MHC II a and b subunits can mix and match to
produce more than 14 different combinations.
5 pts.
2) Explain the why one antibody can only bind one epitope
but one MHC can bind many different peptides.
- On an antibody, the idiotype
has a specific shape that binds to a particular epitope. Each
shape fits the other perfectly.
On an MHC, the binding groove has certain constraints (length
and anchor residues) but the rest of the peptide can vary, and
even modify the shape of the MCH.
15 pts.
3) Using this diagram as a starting place, draw what has to
happen in order to wind up with a functional immunoglobulin (your
choice of specificity and effector function). You must draw each
step that has to happen from DNA to protein. Keep your diagram
at the level of boxes for exons. Do NOT draw anything at the nucleotide
level for this question. Assume all the transciptional units have
the same orientation in this diagram. Make sure your drawings
take into account all the DNA before and after somatic recombination.

Figure taken from ImmunoBiology by Janeway et al., Forth
Edition.
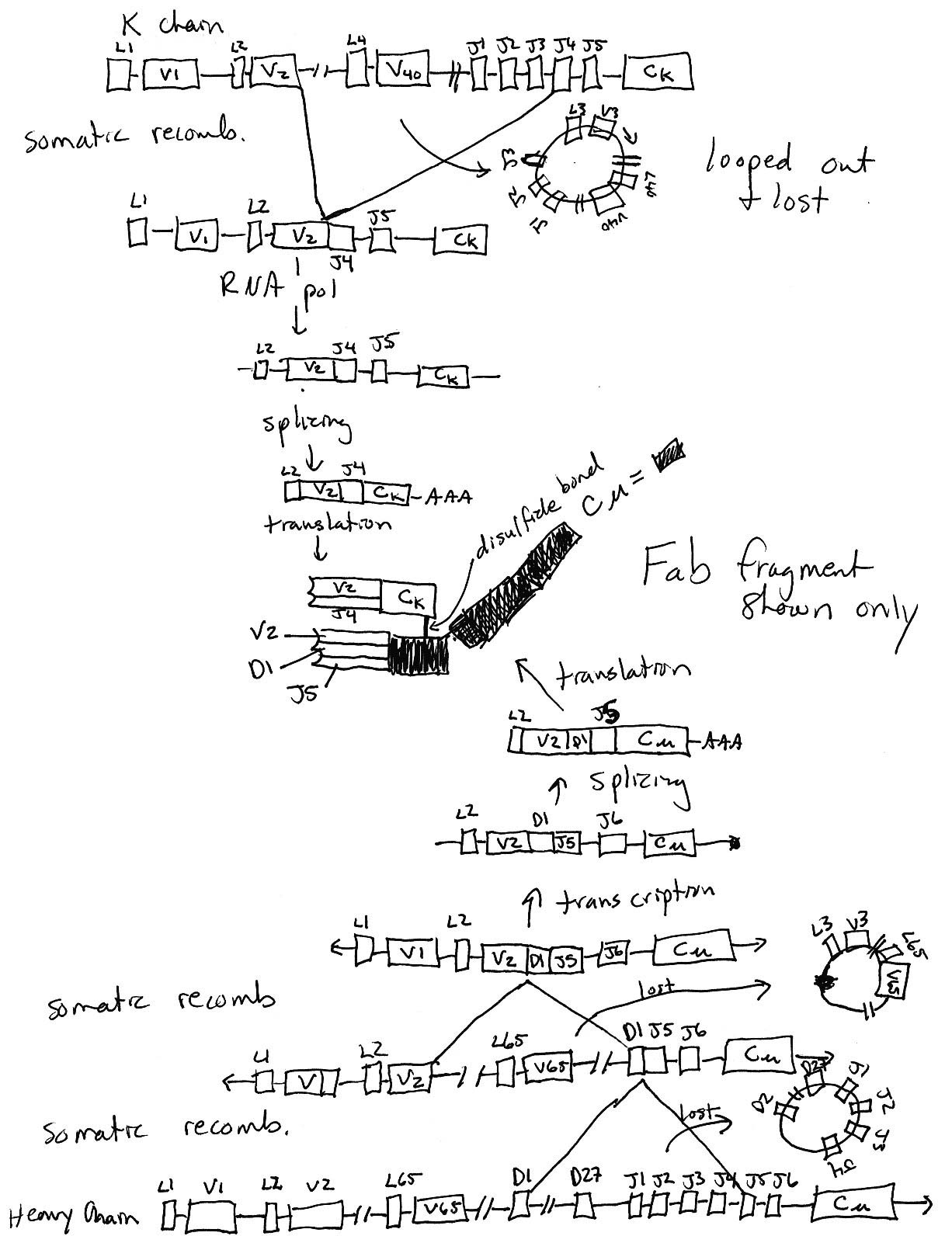 Click on figure to see bigger version.
Click on figure to see bigger version.8 pts.
4) What are complementarity-determining regions? amino acid loops on Ig and TCR that determine the
shape of the ligand that can be bound.
How many are there on a Fab fragment? 6
Then what is the valence of a Fab fragment? 1
On which immunoglobulin domain of a Fab fragment would you find
a complementarity-determining region or regions?- Variable
8 pts.
5) Explain why it is impossible to predict the exact nucleotide
sequence within a coding joint. Your answer should include the
two major factors responsible for this unpredictability. Two main mechanisms:
Coding joint includes P nucleotides which are generated randomly
and thus unpredictably. Variation produced during somatic recombination
when the phosphodiester backbone of nucleotides on one strand
near the hairpin loop is cut so that the nucleotides on this same
strand are flipped over the hairpin "hinge". The exact
number of P nucleotides produced is random and become a part of
the coding joint.
Likewise, N nucleotides are added on randomly by TdT after P nucleotides.
The sequence and number is unpredictable.
8 pts.
6) Is it possible for a B cell to make both membrane bound
IgG and secreted IgG, or is the membrane bound form always IgM
and IgD? Explain your answer.
- Yes, if the B cell is secreting
IgG, then the membrane bound form must also be IgG. In order to
make an IgG, the activated B cell must have undergone isotype
switching which would result in the illimination of the Heavy
chain M/D region. Therefore it is impossible to produce IgG and
IgM at the same time.
8 pts.
7) Explain what aspect or aspects of the immune system is/are
working under these two different conditions:
a. superantigen toxicity- This
antigen is not processed and presented by MHC II, but rather binds
to the more constant portions of MHCII and TCR. It does not see
or interact with the peptide presented by the APC. It stimulates
many CD4+ cells in a non-specific response to secrete cytokine
and a non-productive immune response that can lead to death.
b. allogeneic organ transplant rejection- 5- 10% of the T cells respond to the foreign tissue.
The exact mechanism is not clear but it is either due to strong
binding of the peptide despite the unrecognized MHC allotypes,
or strong binding to the allotypic MHC despite the lack of recognition
of peptide.
12 pts.
8) As you may remember, CD3 is a marker for T cells and CD19
is a marker for B cells. A ficol gradient was used to purify lymphocytes.
a. In general terms, how were the data below produced?- By flow cytometry; all lymphocytes
were labeled with antibodies of two different colors (e.g. anti-CD3
= green and anti-CD19 = red) and measured one by one. Each dot
represents a different cell.
b. Interpret these data fully. Tell me what they mean.- Upper left indicates B cell, Upper
right is non-existant CD3+/CD19+ cell, Lower left is NK cell,
Lower right is T cell. More lymphocytes were measured on the left
than right.
c. What would your conclusion be for the person on the left and
right?- The person on the left
is wt, and has more T cells than any other lymphocyte in circulation.
The perons on the right lacks B cells but NK and T cells seem
OK.
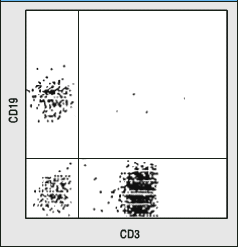
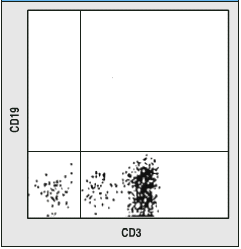
Figures taken from Case Studies in ImmunoBiology by Rosen and Geha, Second Edition.
Return To Immunology Main Page
Return to Immunology Reading Schedule
Go to Biology Course Materials
© Copyright 2000 Department of Biology,
Davidson College, Davidson, NC 28036
Send comments, questions, and suggestions to: macampbell@davidson.edu