
This web page was produced as an assignment for an undergraduate course at
Davidson College. For more information on this exciting new method please refer to the author's link at the bottom of the page
Methods

| Introduction
Proteins are highly involved in cellular function. Many proteins act by interacting with other proteins.(1,8,9,10) Molecular biologists are begining to sequence many different proteins, but their function is often times unknown.(3) The function of these newly discovered proteins can be investigated by studying the relationship that they share with other known proteins.(3) The method that I will be describing is a new application of a decade-old genetic technique that will enable molecular biologists to identify multiple protein-protein interactions.(2,3,4,5,7) This technique can be applied to several areas of research including: novel protein-protein interactions, proteins that interfere or enhance normal protein function, and potential protein-protein binding mutagens.(2,3,4,5,7) Generally, two-hybrid systems are used to discover the function of a known protein by studying other proteins that interact with it.(3,4,5) Sequencing can then be made by screening a cDNA library.(3,4,5) The original methods have recently been improved upon by the addition of multiple reporter constructs, and by using a reverse system to test for potential interference of protein function.(5,8,9) One advantage of this new method is that interaction of the bait protein with an unknown prey protein can reveal the cDNA of the interacting protein, while using huge numbers of potential protein cDNA candidates.(5) By using multiple homologous bait proteins (in one yeast cell), direct comparisons of binding partners within a family of related proteins is possible. Grossel et al.'s method allows one to screen for interactions between prey proteins and two different bait proteins by detecting bait-specific reporters.(5) |
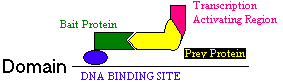
Figure 1. This is a model of transcriptional activation showing how the Bait protein is linked with the transcription activating region so that as the bait protein and the prey protein enter into close proximity, transcriptional activity can take place. This figure was adapted in part from Fields & Song 1989.
The Basics of a Yeast Two-Hybrid System
First, a yeast strain is constructed so that it contains two reporter genes, HIS3 and lacZ, that are dependant upon GAL4 binding sites in their promoters. Also integrated into the yeast strain is a third reporter gene, URA3, that is dependent on LEXA binding sites in the promoter region (See fig. 2).(5) The GAL4 binding sites are able to fuse with a cDNA library-encoded protein attached to a trans-activation domain (TAD). This causes an activation of the two GAL4-dependent reporter genes. Likewise, the LEX binding site is able to fuse with a bait that is attached to a LexA DNA-binding domain. The URA3 reporter gene is activated when it interacts with the prey-TAD fusion protein.(5)
Second, Grossel et al. introduced plasmids that encoded fusions of cdk6 and cdk4 cDNAs with the genes for Gal4 or LexA DNA-binding domains.(5,6) The yeast strain HW18 is then transformed with zeo, elu2, and trp1 plasmids and selected for on SC-L-T (Leu) plates, -T(Trp), +zeo (300µg/ml) plates. Positive growth selections that use a prototrophic selectable marker as reporter genes facilitate the detection of protein-protein interactions.(8) Finally, HW18 is tested for activation of the three reporter genes.(5)
Figure 2. This shows the differential yeast strain HW18 with the HIS3 and lacZ reporter genes as dependent upon the GAL4 binding sites. The URA3 reporter gene is also seen in the figure as being dependant upon the LEX binding site. Figure adapted from Grossel et al. 1999
Library Screening
First, a bait plasmid is constructed from the gene that you are interested in as well as the DNA binding domain of either Gal4 or LexA.(2,5,10) Next, the bait must be transformed with H18 lacking the promoter for the reporter genes and those yeast cells must be selected for.(5) After this is done, you must transform the yeast again with the library plasmids of your known cDNA.(5)Conclusions
In theory, this method could be useful in treating diseases attributed to particular protein-protein or DNA-protein interaction.(8) Using this efficient technique, we now have the ability to screen two proteins simultaneously and identify proteins that bind to either or both bait proteins.(5) The differential two-hybrid yeast is also important in mutagenesis studies by identifying potential chemicals that could disrupt certain cellular functions that involve protein-protein interaction, while sparing other cell functions.(2,5) The rate that new proteins are being identified is incredible. The Human Genome Project has contributed many new protein sequences that still have unexplored functions. For this reason alone, the yeast two-hybrid system for discerning differential interactions using multiple baits will be an exciting new tool for molecular biologists everywhere. (3,4,5)
| 1. Brent, R, Ptashne, M. 1985. A eukaryotic transcriptional activator
bearing the DNA specificity of a prokaryotic repressor. Cell 43:729-36
2. Fields, S, Song. OK. 1989. A novel genetic system to detect protein-protein interactions. Nature 340:245-46 3. Finley, Russel. Brent, Roger. Two-hybrid analysis of genetic regulatory networks. <http://cmmg.biosci.wayne.edu/finlab/YTHnetworks.html> Accessed 2000 Feb 18 4. Finley, R. 1997. Examining the function of proteins and protein netwlrks with the yeast two-hybrid system. <http://cmmg.biosci.wayne.edu/finlab/Update.html> Accessed 2000 Feb 18 5. Grossel, M, Wang, H, Gadea, B, Yeung, W, Hinds, P. 1999. A yeast two-hybrid system for discerning differential interactions using multiple baits. Nat Biotechnol 17:1232-33 6. Gyuris, J, Golemis, E, Chertkov, H. 1993. Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell 75:791-803 7. Mendelsohn, AR, Brent, R. Applications of interaction traps/two hybrid systems to biotechnology research. <http://www.molsci.org/brentlabweb/andyweb/m&b.html> Accessed 2000 Feb 18 8. Vidal, M, Brachmann, RK, Fattaey, A, Harlow, E, Boeke, JD. 1996. Reverse two-hybrid and one-hybrid systems to detect dissociation of protein-protein and DNA-protein interactions. Proc Natl Acad Sci 93:10315-20 9. Vidal, M, Braun, P, Boeke, JD, Harlow, E. 1996. Genetic characterization of a mammalian protein-protein interaction domain by using a yeast reverse two-hybrid system. Proc Natl Acad Sci 93:10321-26 10. Xu, WC, Mendelsohn, AR, Brent, R. 1997. Cells that register logical relationships among proteins. Proc Natl Acad Sci 94:12473-78 |
Now you can return to my
Molecular Biology Home Page
Return to Davidson College Biology Department Home Page
© Copyright 2000 Department of Biology, Davidson
College, Davidson, NC 28036
Send comments, questions, and suggestions to:
ankazama@davidson.edu