MacDNAsis Analysis of Hexokinase Type 1
The amino acid sequences of hexokinase 1 from the following five species were compared using MacDNAsis software.
The source for these sequences is GenBankThe results of the MacDNAsis anaylsis of hexokinase 1 are shown below.
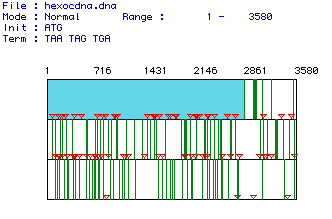
Figure 1 shows the largest open reading frame for the hexokinase mRNA
sequence from Homo sapiens. The red lines indicate the location
of start codons and the green lines indicate the location of stop codons.
The open reading frame is shown in light blue and extends from nucleotide
1 through nucleotide 2835. There are 3580 nucleotides in this mRNA
sequence overall.

Figure 1. The largest open reading frame for the hexokinase gene is shown in blue. This ORF was generated by analyzing the mRNA sequence from Homo sapiens
Using the open reading frame of Homo
sapiens shown in Figure 1, the translation sequence of the protein
was determined. Figure 2 shows the amino acid content of the translated
hexokinase protein. The open reading frame of human hexokinase 1
mRNA encodes a protein consisting of 945 amino acids. The molecular
weight of the protein is 105359.71. The percentage of each amino
acid is listed in the graph. Notice that every amino acid is present
in some quantity in hexokinase.
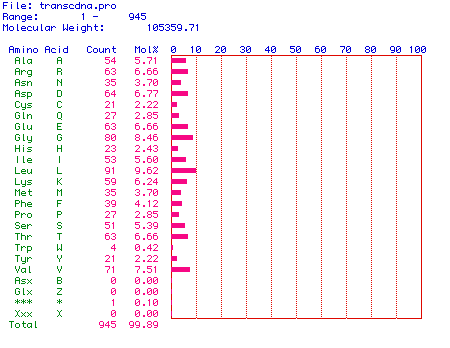
Figure 2. Amino acid content of human hexokinase 1.
In order to determine the cellular location of hexokinase 1, a Kyte
& Dolittle hydropathy plot of the human protein was generated.
This hydropathy plot is shown in Figure 3. Hydrophobic domains are
positive values and hydrophilic domains are negative values. A threshold
line was placed on the value of +2 in order to clearly delineate the most
hydrophobic values from less hydropbobic values. Several domains
exhibit hydrophobicity values greater than +2, indicating that hexokinase
1 could be an integral membrane protein in humans.
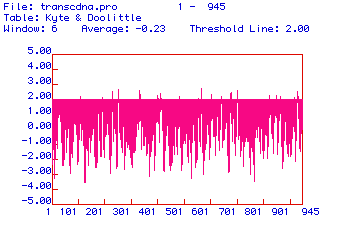
Figure 3. Hydropathy plot of the amino acid sequence of human hexokinase 1 with a threshold line set at +2. Hydrophobic domains are positive and hydrophilic domains are negative.
The secondary structure of human hexokinase 1 is further elucidated
in the antigenicity plot shown in Figure 4. Hydrophilic domains are
positive and hydrophobic domains are negative. Since the most hydrophilic
domains are the portions of the protein that extend farthest out from its
center, these domains are the most likely candidates for effective epitopes.
Two domains of hexokinase 1 are distinctly hydrophilic: amino acids ~210-220
and amino acids ~700-710. These peptide sequences would be good epitopes
against which to generate monoclonal antibodies for human hexokinase 1.
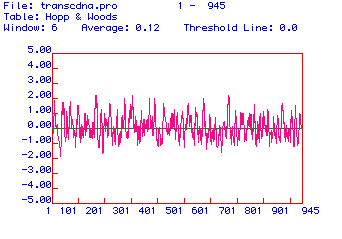
Figure 4. Antigenicity plot of the amino acid sequence of human hexokinase 1. Positive values are hydrophilic and negative values are hydrophobic.
A prediction of the secondary structure of human hexokinase 1 is
shown in Figure 5. Blue segments are alpha-helices, red segments
are pleated sheets, green segments are turns, and black segments are coils.
Because the amino acid sequence turns in several locations, this secondary
structure prediction further suggests that human hexokinase 1 could be
an integral membrane protein.
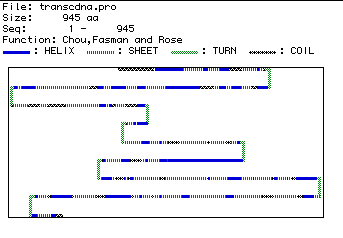
Figure 5. Predicted secondary structure of human hexokinase 1, showing regions of structural difference.
The amino acid sequences of hexokinase 1 of five different species
were compared to determine homology. This multiple sequence alignment
is shown in Figure 6. The sequences used to produce this figure are
those listed at the top of the page. Homologous amino acids are highlighted
in black, and dashes indicate spaces that were inserted to maximize alignment.
The multiple sequence alignment was generated for the entire amino acid
sequence of the five species, and a representative portion is shown here,
consisting of amino acids 1-350 of the multiple alignment. Note that
the carboxyl terminus of hexokinase has diverged between the species, but
that the more interior regious have retained greater homology. Also
note the greater homology between the human, mouse, and rat sequences,
and the divergence of homology between the yeast and Arabidopsis
sequences.
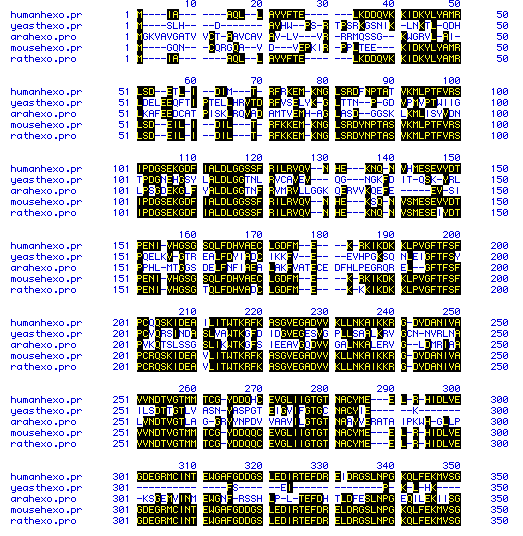
Figure 6. Multiple sequence alignment of the amino acid sequence of hexokinase 1 from five species. Homologous sequences are in black and dashes indicate spaces that were inserted to maximize alignment.
The greater homology between the human, mouse, and rat hexokinase
sequences is confirmed in the phylogenetic tree shown in Figure 7.
The mouse and rat hexokinase sequences are 94.2% homologous, and the human
hexokinase sequence is 90.4% homologous to the rodent sequences.
The divergence in sequence of the yeast and Arabidopsis hexokinase
is indicated by their much lower homology: 17.6% for the yeast and 16.7%
for the Arabidopsis. The similarity between the three mammalian
sequences is to be expected, and the lack of homology of the other two
species indicates the much greater evolutionary divergence of these species.
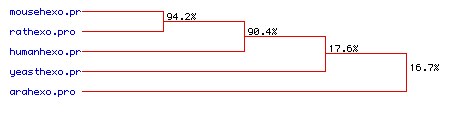
Figure 7. Phylogenetic tree indicating the amino acid sequence homology of hexokinase 1 for five species.
All figures and data on this page were generated using MacDNAsis software.
Davidson College Molecular Biology Home Page
Return to my Molecular Biology Main Page
Contact me: miosgood@davidson.edu
This web page was produced as an assignment for an undergraduate course at Davidson College.
Copyright Davidson College 2000