Voyager is a new method in molecular biology which expedites the process of recombination of proteins. It is more advantageous to molecular biologists as a method because it is fast and easy and does not require complete precision (for some parts). The method employs the Herpes Simplex Virus VP22 protein as the vector plasmid utilized in the transport of a specific protein to particular target cells. This Herpes Virus is able to translocate between cultured cells, and this ability is important to recombination because VP22 transports the specific protein of interest into the nucleus of almost every cell for more efficient transfection. It is this efficiency which accelerates the process.
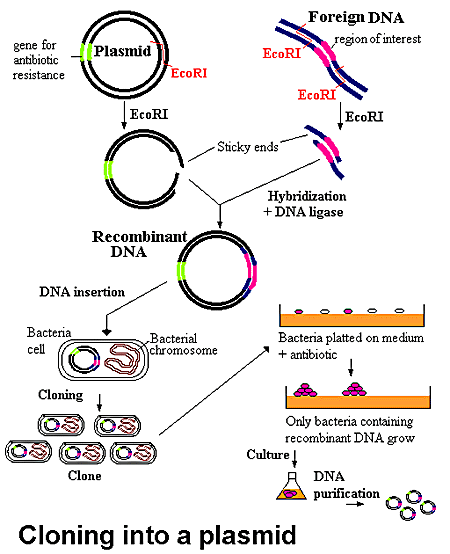
In recombination, a plasmid is used as a transport for a specific DNA sequence. First, the plasmid and the chosen DNA are digested with a particular restriction enzyme to produce sticky ends which can then base pair when the different fragments are mixed together. The plasmid must also contain several other visibly recognizable genes including antibiotic resistance, such as to ampicillin, and the lacZ gene which encodes the production of ß-galactosidase in order to ascertain which plasmids took up the DNA of interest. With the mixing of the plasmids and the specific DNA, the DNA sequence becomes integrated into the plasmid by the base pairing of the sticky ends. DNA ligase must then be added in order to produce covalent bonds between the two parts. If the plasmid took up the chosen DNA in the middle of a visibly operating gene (such as the lacZ gene), then the plasmid will lose the functionality of the lacZ gene. This can be seen when the plasmids are put into bacteria which lack the appropriate gene, such as the lacZ gene, by transformation. The cells are then plated in a medium which contains whatever antibiotic the plasmid is resistant to, such as ampicillin. In the case of ß-galactosidase, a specific sugar called X-gal is cleaved by the enzyme and gives off a blue end product. Therefore, if the colony turns blue instead of remaining white, it should be disregarded because it does not necessarily contain the DNA of interest. The white colonies result from bacteria which integrated the plasmid and chosen DNA. These growths can then be cloned and delivered to the target cells.

This particular figure uses EcoR1 as the restriction enzyme. As you can see, the plasmid contains antibiotic resistance which is essential in the identification of the correct colonies.
The vectors which Invitrogen has designed are pVP22/myc-His and pVP22/myc-His-TOPO. The first vector is especially useful because it has eight distinct restriction sites which add to the efficiency of the method and make for advantageous cloning. The second vector carries the TOPO technology which concedes 5 minute cloning of PCR products with Taq amplification. There are also various other features which make these admirable vectors. When a cell is transfected with a vector containing VP22, the cell becomes more localized in the cytoplasm.
Several steps are involved in the production of a homogeneous population with the VP22-fusion protein in the nucleus:
Campbell, Neil A. Biology. 4th Ed. New York: The Benjamin/Cummings Publishing Company, Inc., 1996.
Tue, Dec 8, 1998 9:15:09 PM GMT. Plasmid Insertion. Access Excellence Genentech. <http://www.gene.com/AE/AB/GG/plasmid.html> Accessed 1999 February 13.
Fri, Jan 22, 1999 8:27:11 PM GMT. Invitrogen Web Resource-Home Page.
<http://www.invitrogen.com/> Accessed 1999 February 3.
![]()
e-mail me at: alcaldwell@davidson.edu