My Favorite Yeast Expression(s):
Ste2 (annotated) and YFL030W
(unannotated)
This web page was produced as an assignment
for an undergraduate course at Davidson College.
My Favorite Annotated Yeast Gene's
Expression: Ste2
By mining the databases available,
the protein expression given by the function of the gene Ste2 may be further
analyzed. Function
Junction and Expression
Connection, searches of many different databases performed on the SGD
database, initiates the discovery of information about this gene.
The Function Junction website's use of the
Yeast Microarray Global Viewer (provided by the Laboratoire de genetique moleculaire,
Paris, France) is an excellent starting place for exploring this gene's expression.
A couple experiments found here are of note:

Screen shots from the Function
Junction website. Click on each to see an enlarged, more detailed view.
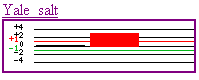
First, the Yale experiment shows
some interesting findings. Of the three different types of conditions the
yeast were grown (1M NaCl for 10, 30, and 90 minutes) the Ste2 gene was induced
greatly during the the 30 minute condition. This would imply that there is
some optimal time for the presence of salt that somehow causes the type a
yeast cell to increase its production of Ste2 (encoding alpha pheromone receptor),
presumably for reproduction.
The Hardwick rapamycin experiment
also provides some interesting information. When rapamycin is added to the
yeast cell, induction of Ste2 occurs. Even more interestingly, when amino
acid starvation and addition of rapamycin is the experimental condition, induction
of the Ste2 gene increases dramatically. This can be interpretated as the
cell's attempt to reproduce at times of high stress, which may be counterintuitive.
Unfortunately, no genes were
found to have strong interactions with STE2, as is indicated on this rather
depressing Function Junction graphic:
Screen shots from the Function
Junction website. Courtesy SGD
(2003).
The Expression Connection yielded
some important hits, which show genes with similar expression patterns, therby
making them "guilty by association" and, thus, related to one another.
The most obvious experimental scenario where one would expect an induction
of the alpha pheromone receptor would be when alpha factor is introduced to
the cell. Indeed, this is the case:
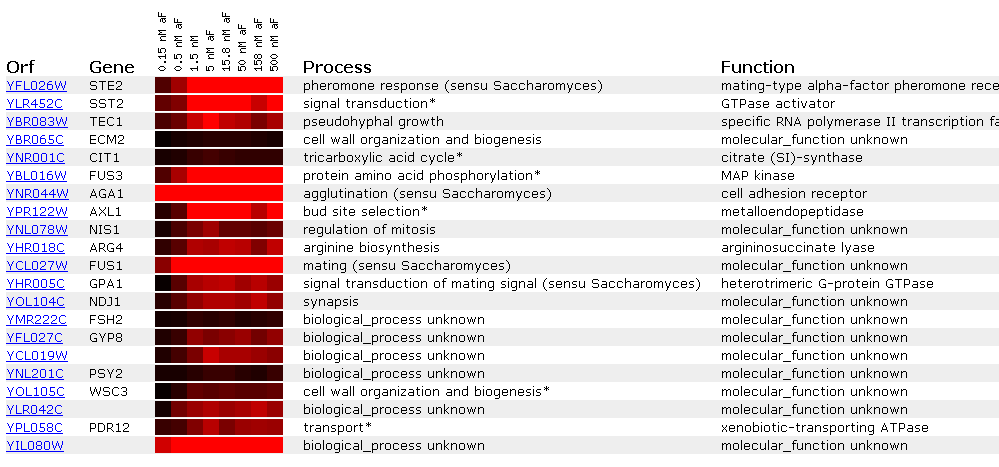
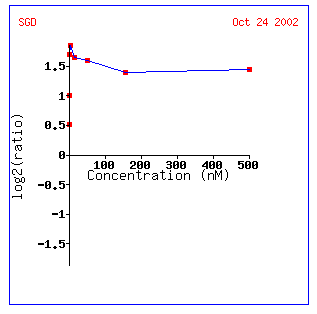
Expression at different
alpha-factor concentrations for STE2
Screen shots from the Expression
Connection website. Courtesy SGD
(2003).
As expected, Ste2 is induced when alpha factor
is added to the yeast cell. Interestingly enough, however, it doesn't really
matter how much alpha factor is added after 1.5 nM. This search also shows
several other yeast genes associated with mating: FUS1, FUS3, GPA1, and AXL1.
Although Function Junction didn't find these genes directly interacting with
STE2, it would not be surprising if these genes were found in to be in some
connection, remote though it may be.
Expression in time response to
alpha-factor for STE2
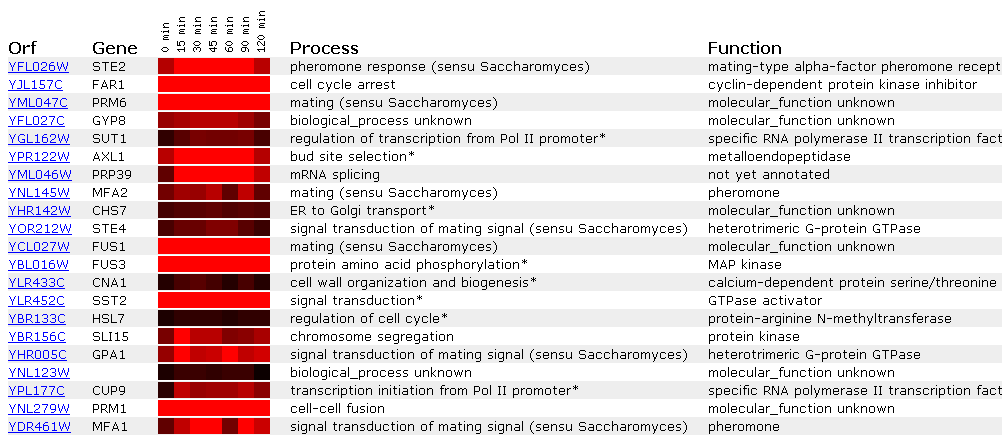
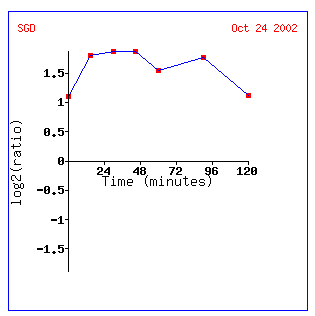
Screen shots from the Expression
Connection website. Courtesy SGD.
An experiment taking the yeast genomes at different
times after the alpha factor was added yielded similar results. Again, production
of alpha pheromone receptors when alpha pheromone is present is expected,
but this data also suggests that there may be some upper limit (120 minutes?)
when the cell may acclimate itself to the condition of extra alpha factor
availability or the cell may not have the available material to produce more
alpha pheromone receptors.
Expression in response to environmental
changes for STE2
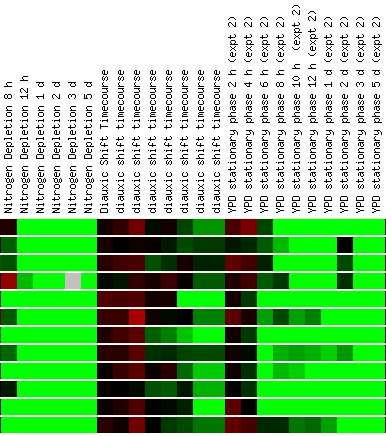
Screen shots from the Expression
Connection website. Courtesy SGD.
Also notable are those experiments that related
the activity of Ste2 to environmental factors, like those that yeast cells
may encounter outside of the laboratory. As would be expected, mating factor
receptors is repressed when the cell is under large environmental stress.
This is the case above, when the cell is attempting to grow in an environment
that is depleted of nitrogen. Also, repression of Ste2 can be seen when the
cell is in the stationary phase upwards of 6 hours. Again, this is expected,
as pheromone receptors are not needed when there is no pheromone for the cell
to uptake to initiate reproduction. As would be expected a gene under a similar
name (Ste6) is very similar in expression to Ste2. This gene is involved in
a factor transport, which is why one would name it in connection
with its cousin, Ste2. a factor is produced in greater amounts
in the cell when other cells are present to begin sexual reproduction. This
makes sense here -- both the genes for alpha factor receptors and a
factor transporters are induced when alpha factor is present in the cells.
The other experiments that the Expression Connection
website looks for (response to DNA-damaging agents, expression during the
diauxic shift, evolution of expression during glucose limitation, regulation
by the PHO pathway, expression during the cell cycle, response to histone
depletion, expression during sporulation, and response to varying zinc levels)
yielded fairly uninteresting results. Overall, the gene Ste2 seems fairly
cut and dried. The cell cycle view confirms that the seven-transmembrane region
protein is being made constantly for use in the membrane of the Sacchromyces
cell. This production increases when the factor that binds to it (alpha pheromone)
is present. The yeast cell halts this process when it is under heavy stress
and is more concerned with survival than reproduction, or is in a stationary
phase until it comes into contact with another yeast cell to mate with.
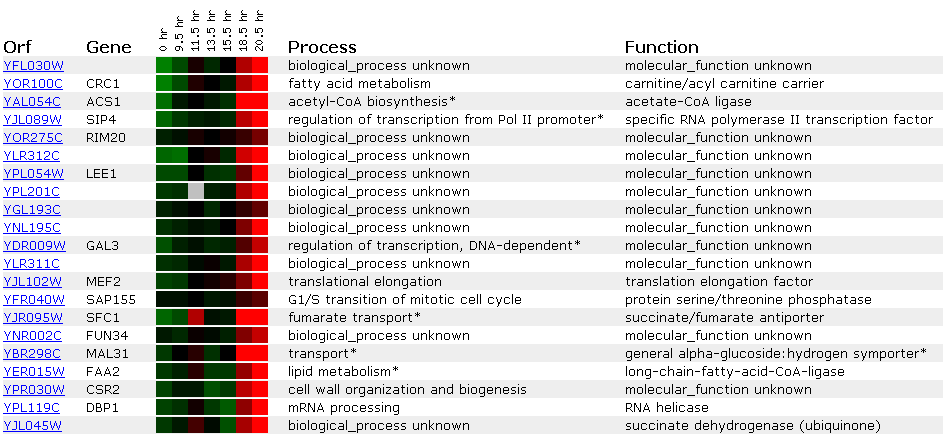
My Favorite Unannotated Yeast Gene's
Expression: YFL030W
The expression of the unannotated
yeast gene is more intriguing, as its function is unknown according to the
SGD database.
Unfortunately, but as expected, no genes were found to be connected with the
unannotated gene:
Screen shots from the Function
Junction website. Courtesy SGD
(2003).
Again, simarly to the Ste2 gene's hits on the
Function Junction database, there were some experiments involving YFL030W
that stuck out:

Screen shots from the Function
Junction website. Click on each to see an enlarged, more detailed view.
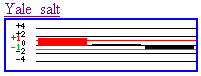
Several predictions about the function of the
unannotated gene can be made from the preceding experiments. First, according
to the Yale salt experiment, it appears NaCl, after a long period of time
(90 minutes), induces the production of this gene (graph above goes from 90
to 30 to 10 minutes, left to right). Whether this is due to experimental error
or is an actual effect of the gene product is uncertain, however. Also, many
different processes could be affected by salt, so the interpretations from
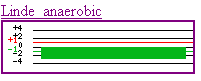
this data are boundless. The second experimental condition (Linde
anaerobic) was growing the cell in anaerobic conditions. This produced
repressed expression of the gene. A hypothesis can be made that this gene
is only produced in aerobic conditions or has is a gene where oxygen is necessary
to complete its function. Finally, the Jelinsky
experiment added MMS, an alkylating agent, to the yeast cells. In our gene,
this produced a large induction of the gene product, maybe indicating that
the product is needed to help repair the cell when DNA-damaging agents are
added.
The Expression Connection webpages also yielded
some interesting findings, although many of the experimental conditions produced
no change in the expression of YFL030W:
Expression during the diauxic
shift for YFL030W
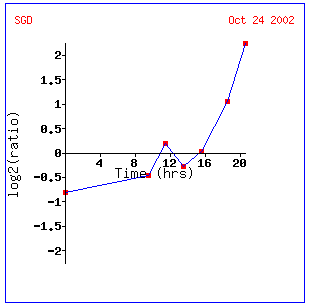
Screen shots from the Expression
Connection website. Courtesy SGD
(2003).
There is a fairly interesting pattern here, where
immediate repression eventually yields to induction in an almost exponential
fashion. This experiment indicates again that this gene may be involved in
anaerobic respiration, as diauxic shift is a change from anaerobic to aerobic
respiration. In addition, this appears to be in conjunction with the Linde
article -- anaerobic conditions caused a repression of YFL030W. In the experiment
above, anaerobic conditions show repression. However, as aerobic respiration
becomes available, induction of the YFL030W gene is seen. The genes that were
found to be similar to our unannotated gene are fairly vaired. SFC1 is of
particular note, as it is also found in a different experiment, further connecting
it with YFL030W. This gene's biological process is fumarate transportation,
which is essential for growth on ethanol or acetate (SGD).
How this connects with the oxygen dependancy of this gene remains to be seen.
Expression in response
to environmental changes for YFL030W

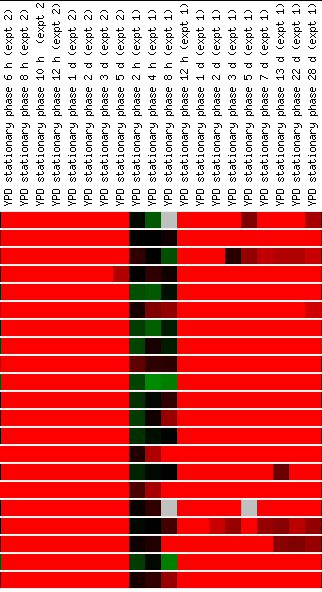
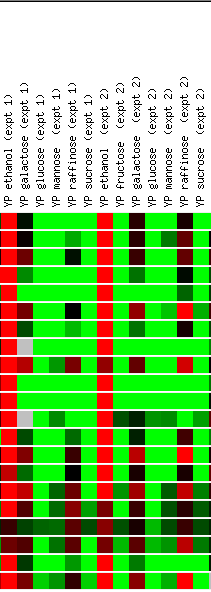
Screen shots from the Expression
Connection website. Courtesy SGD
(2003).
The results of the environmental changes experiment
is very informative, as the outcomes are very strong (both induction and repression),
and since it involves so many conditions, those genes that "fit"
well are more likely to make the gene in question "guilty by association".
YFL030W is induced during heat shock, during the yeast cell's stationary phase
(especially after half a day), and when ethanol is added to the media. When
sugars are added (glucose, mannose, and sucrose especially) the gene is dramatically
repressed. Again, these data indicate YFL030W's importance during aerobic
cellular activity. The repression in the presence of sugars in perplexing,
however, as it is hard to see a connection between sugar addition and respiration.
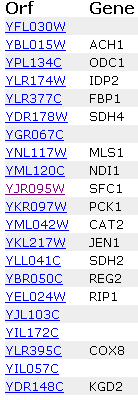

Screen shots from the Expression
Connection website. Courtesy SGD
(2003).
The genes listed above identify with YFL030W
most uniquely. This is again a variety of gene products, but fumarate transport
(SFC1), acetate metabolism (ACH1), and aerobic respiration (RIP1) all seem
like viable options for this gene product of the unannotated ORF. The other
experiments weren't extremely helpful. The cell cycle analysis, for example,
showed that the gene is held fairly constant throughout the cell cycle. It
is fairly clear that the gene comes to play in extreme conditions. In any
case, it is almost certain that anaerobic and aerobic conditions hold the
key to the product of the unannotated gene on chromosome 6.
The data gleaned from the original
look at YFL030W (without microarray data) was not very informative, and
not much could be hypothesized. However, this data surely preliminarily confines
the unannotated gene to a more selective group.
References:
Jelinsky,
Scott A. and Leona D. Samson. Global response of Saccharomyces
cerevisiae to an alkylating agent. PNAS, Vol. 96, Issue 4, 1486-1491,
February 16, 1999
Linde,
J. J. M. ter. Genome-Wide Transcriptional Analysis of Aerobic and Anaerobic
Chemostat Cultures of Saccharomyces cerevisiae. Journal of Bacteriology,
December 1999, p. 7409-7413, Vol. 181, No. 24.
Palmieri L, et al. (1997) Identification of the yeast ACR1 gene
product as a succinate-fumarate transporter essential for growth on ethanol
or acetate. FEBS Lett 417(1):114-8
Saccharomyces
Genome Database
Return to Dan Pierce's Genomics Web Page
Genomics
Biology
Home Page


© Copyright 2003 Department of Biology, Davidson College, Davidson,
NC 28035
Send comments, questions, and suggestions to: dapierce@davidson.edu




















