This page was produced as an assignment for an undergraduate course at Davidson College.
How to do a BLAST2
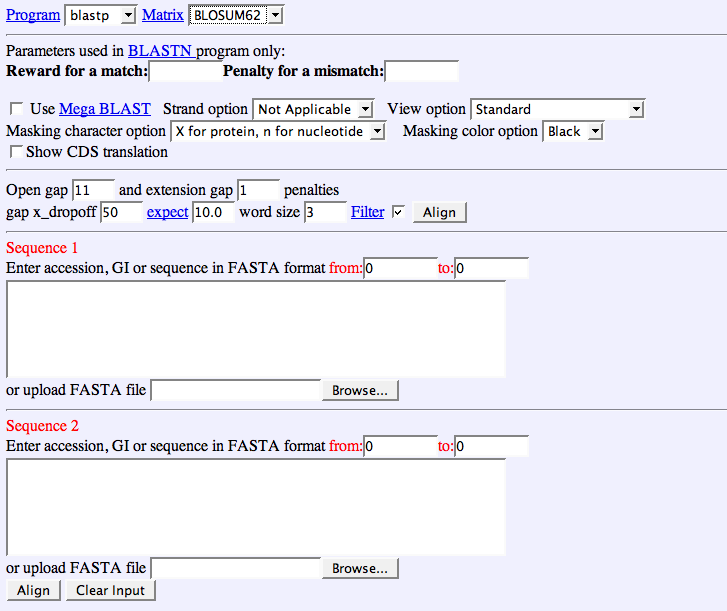
Clicking the BLAST2 link above will bring you to the BLAST2 page, which looks like this:

On the "Program" line, you can choose blastn for blasting two nucleotide sequences, or blastp for blasting two amino acid sequences. If you pick blastp, you can select which scoring matrix you want to use. For mor information on scoring matrices, see "About BLAST algorithms" below. If you choose blastn, you can pick your reward for a match (the default is +1) and your penalty for a mismatch (the default is -3). You can also change your gap opening and gap extension penalties. You will want high gap opening penalties if you expect that your query sequence may form a protein, because one gap will de-align the entire amino acid chain. After setting all of your parameters (the defaults usually work well), enter your sequences in the two boxes, or upload them using the "browse" option. Click align.
The results page will look something like this:
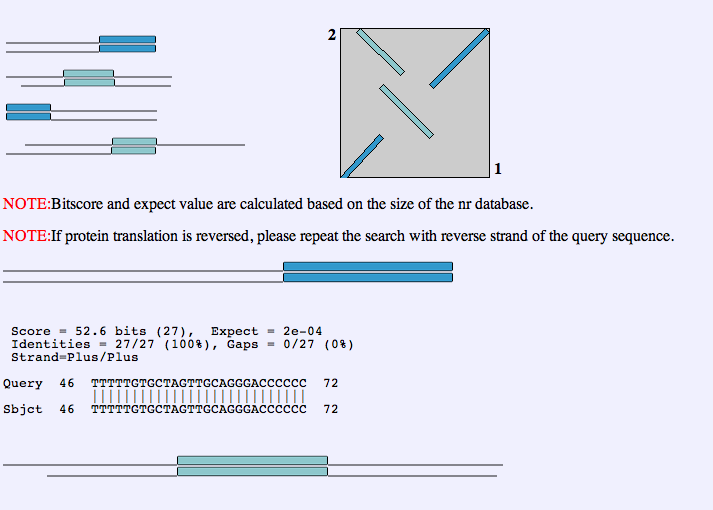
The graph is a dot plot and it shows the user visually how the entirety of the two strands aligned. The blue diagonal lines with a slope of +1 indicate that the beginning and end of the two sequences aligned. The light blue line with a slope of -1 in the center indicates that in the middle of the two sequences, there was an inversion. The light blue line in the top left corner indicate an inverted repeat from the beginning of sequence 1 at the end of sequence 2. Below the dot plot, each alignment is evaluated separately.
How to use BLAST Front Page
Genomics Page
Biology Home Page
Samantha's Home Page
Halorhabdus utahensis Genome Wiki