This web page was produced as an assignment for an undergraduate
course at Davidson College.
GPA1 and its Non-annotated Neighbor YHR003C.
GPA1 is a well annotated gene located on Saccharomyces cerevisiae chromosome 8. It is known to be the alpha subunit of a heterotrimeric G protein that is involved in the mating signal transduction pathway. While the function of GPA1 is well known, its nearby neighbor YHR003c is a hypothetical ORF whose cellular component, molecular function, and biological process are unknown. (Dolinski et. al., 2003).
Chromosomal Location
Both the annotated GPA1 and the non-annotated YHR003C are located within a small region of Saccharomyces cerevisiae chromosome 8 as shown below:
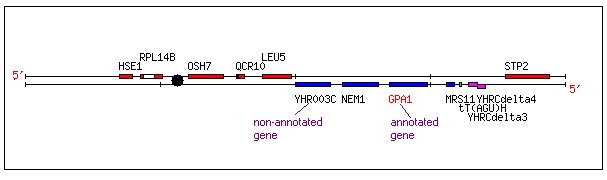
Figure 1. Chromosomal Location of YHR003C and GPA1. This image shows the location of YHR003C and GPA1 on the crick strand of yeast chromosome 8 (coordinates 100000 to 120000). Image from: http://db.yeastgenome.org/cgi-bin/SGD/ORFMAP/ORFmap?seq=YHR005C (Cherry, et. al., 1997). Permission Pending.
GPA1 Basic Gene Information
Saccharomyces Genome Database: <http://db.yeastgenome.org/cgi-bin/SGD/locus.pl?locus=YHR005C>
A large amount of information about the gene GPA1 as well as its protein product can be found at the Saccharomyces Genome Database (SGD). Included is the information that this gene is located on the crick strand of chromosome 8 and spans 1419 bases (from 114910 to 113492).
Genbank:
NC_001140 - The nucleotide sequence for GPA1 can be found in the entry for yeast chromosome 8 at this accession number.
NP_011868- The corresponding amino acid sequence for GPA1 is located at this accession number. This page states that the encoded protein of this gene is 472 amino acids long.
SwissProt: <http://us.expasy.org/cgi-bin/niceprot.pl?P08539>
The information contained on this website about GPA1 states that the protein product of this gene belongs to the G-alpha subfamily 3, but notes that the gene contains an insertion (corresponding to 109 residues) that does not appear in the alpha subunits of G proteins in other species.
The Role of GPA1 in the Yeast Mating Pheromone Response
Saccharomyces cerevisiae (baker's or budding yeast) is a unicellular eukaryotic organism. The individual cells are diploid and belong to one of the two mating types "a" or "alpha". They can reproduce asexually by budding or sexually by fusing with a cell of the opposite mating type. Each cell secretes a mating pheromone that can bind to receptors on the opposite mating type. When a cell detects a pheromone secreted by the opposite mating type, a signaling cascade occurs with results in cellular differentiation, cell cycle arrest and the formation of an oblong cell shape called a "shmoo." Two shmoos of opposite mating types can then bind together and fuse to form a diploid cell (Blackwell, et. al. 2003).
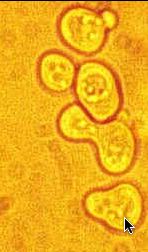
Figure 2. Microscope Image of Shmoos. This image shows the typical oblong shmoo shape of the yeast cells that have been exposed to mating pheromone. Image from: www.phys.ksu.edu/gene/photos/lc.html Permission Pending.
The basic signaling pathway from the pheromone stimulus to the altered gene transcription in the nucleus is shown below:
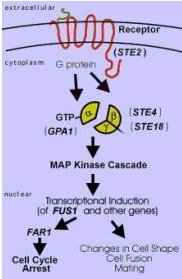
Figure 2. Mating Signal Transduction Pathway. This image shows a basic flow diagram of the signaling pathway from the reception of a mating signal pheromone by the STE2 receptor at the plasma membrane to the transcriptional changes that lead to cell differentiation. GPA1 is shown as the alpha subunit of the G protein. Image from: http://dbb.urmc.rochester.edu/bcbp/members/faculty/Dumont_Mark.HTMl Permission Pending.
According to the SGD, the protein product of the GPA1 gene plays a role in the mating signal transduction pathway in both "a" and "alpha" cells. The role of this gene in yeast mating, as described in Gene Ontology terms, is detailed below:
Molecular Function
GPA1 is the alpha subunit of the heterotrimeric G-protein which is coupled to the mating pheromone receptor. The molecular function of this subunit is to perform catalysis of the hydrolysis of GTP to GDP and water. In this function, GPA1 is similar to many proteins which are involved in stimulating or inhibiting pathways involving ion channels. <http://db.yeastgenome.org/cgi-bin/SGD/GO/go.pl?goid=3927>
Biological Process
The Gene Ontology website cites the biological process of GPA1 as its participation in signal transduction during mating that results in cellular fusion. <http://db.yeastgenome.org/cgi-bin/SGD/GO/go.pl?goid=750> Originally, it was only known that GPA1 played some role in either transmitting, inhibiting, or amplifying the mating signal, but recent work has helped to clarify the precise role of GPA1 in this pathway. SwissProt <http://us.expasy.org/cgi-bin/niceprot.pl?P08539> gives more detailed information by indicating that GPA1 is thought to release the beta and gamma subunits of the heterotrimeric G protein when stimulated by the coupled pheromone receptors STE2 or STE3. In this way, GPA1 plays a role in stimulating the pheromone response, since releasing the beta and gamma subunits allows them to target other proteins and begin the pheromone response signaling cascade (Swissprot, 2003). Recent research suggests that in addition to releasing these subunits, GPA1 plays a role in inhibiting and downregulating the pheromone response. GPA1 has been found to affect the localization of FUS3, a MAP kinase that is induced during the pheromone response and which plays a role in the cellular differentiation in response to the pheromone. GPA1, in cooperation with other proteins such as MSG5, appears to directly bind to FUS3 and prevent its accumulation in the nucleus. As a downregulator of the pheromone response pathway, GPA1 allows cells to "adapt" to a prolonged pheromone signal and resume the cell cycle if no fusion occurs (Blackwell, et. al., 2003).
Cellular Component
Since GPA1 is part of the heterotrimeric G-protein that is coupled to the integral membrane receptor STE2, it is expected that the protein will be located near the plasma membrane. SGD confirms this expectation, stating that the cellular component for GPA1 is the cell membrane. <http://db.yeastgenome.org/cgi-bin/SGD/GO/go.pl?goid=5886> With the recent research that details the role of GPA1 in affecting FUS3 levels in the nucleus, however, suggests that this protein might also be found elsewhere in the cytoplasm or the nucleus during a mating signal (Blackwell, et. al., 2003).
Protein Interactions
More information about the role of GPA1 in the life of yeast can be obtained by a search of the The GRID database of protein interactions <http://biodata.mshri.on.ca:80/grid/servlet/SearchResults?news=welcome&keywords=GPA1>. Some of the interactions found my affinity precipitation, the two-hybrid method, and other similar methods are expected considering the known function of GPA1 in the mating pathway. GPA1 interacts with STE4 and GPA2, the other two subunits of the heterotrimeric G protein, and STE11, another protein involved in the mating signal cascade. Some of the associated proteins, however, have unknown roles (like YBR116C) or are active in pathways thought to be unrelated to the mating pathway. For example, GAC1 is associated with the glycogen synthase-2 pathway, but was found to interact with GPA1. This indicates that more study of each of these proteins and their potentially numerous cellular roles is needed.
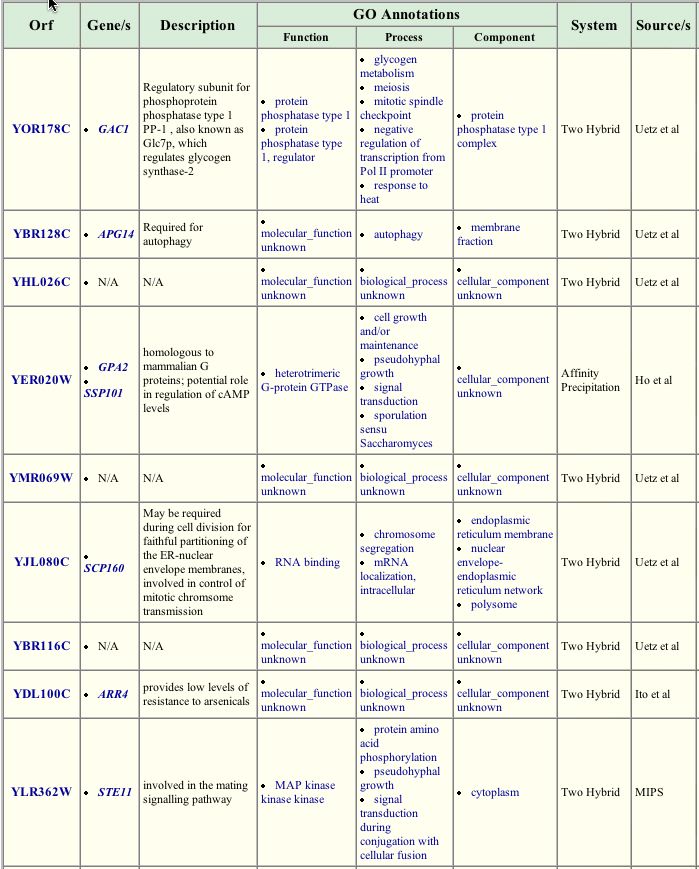
Figure 3. Proteins interacting with GPA1. This table lists some of the proteins found to interact with GPA1. Within the list, some proteins have known functions in the mating pathway, others have known functions elsewhere, and some are not annotated. Image from: http://biodata.mshri.on.ca:80/grid/servlet/SearchResults?news=welcome&keywords=GPA1 Permission Pending.
Polymorphisms and Effects
According to the SGD, cells in which the GPA1 gene was deleted were viable but experienced defects in the pheromone response (Dolinski, et. al., 2003). Other resources outline the effects of specific polymorphisms in yeast cell function. Yvert, et. al. found a strain of yeast with missense mutations in the GPA1 gene that led to two amino acid changes in the protein product. One of these changes (S to I at amino acid number 469) was nonconservative and affected an essential binding site of this protein to the pheromone receptor STE2. The mutation resulted in a defect in the mating pheromone response in these yeast cells, as would be expected from a disruption in the connection between the pheromone receptor and the two G protein subunits which transmit the signal to the rest of the cell. The research cited the fact that point mutations in other nucleotides that affect amino acids 467 and 468 also cause mating response defects. Mutant strains that lacked 22 terminal residues gave a result consistent with the proposed role of GPA1 in inhibiting the mating response. These mutants had growth defects and appeared to experience constitutive activation of the pheromone response. (Yvert, et. al., 2003). Other research has characterized a mutation that creates a hyperactive GPA1 allele (form E364K). Yeast cells with this allele are found to be completely resistant to to pheromone stimulation, again confirming the inhibitory role of GPA1 in the yeast mating response pathway (Li et. al., 1998).
Importance of Study
The investigation of genes like GPA1 and its encoded G protein subunit is very important both for the general understanding of yeast as a model system, and also to increase knowlege about cellular signaling pathways in general. Trimeric G proteins are active in many different signaling pathways across species (Dumont, 2003). Research has also shown that the yeast mating pathway is very similar to the human beta 2 adrenergic signaling pathway (King et. al., 1990). Better understanding of the pheromone signal transduction pathway thus has potential benefit in many areas.
Investigation of YHR003C
In the SGD, the cellular component, biological process, and molecular function are listed as unknown for hypothetical ORF YHR003C. <http://db.yeastgenome.org/cgi-bin/SGD/locus.pl?locus=YHR003C> This site indicates that in a systematic deletion study, yeast with a deletion of this gene were viable, so the protein product of the gene does not appear to have a function essential for basic survival. No articles were found with a PubMed search for YHR003C, confirming that very little study of this gene has been done.
Basic Gene Information
Saccharomyces Genome Database: <http://db.yeastgenome.org/cgi-bin/SGD/locus.pl?locus=YHR003C>
This database indicates that the YHR003C gene is 1289 bp long and is located on the crick strand of chromosome 8 from 111310 - 110021. The genomic DNA sequence can be found at <http://db.yeastgenome.org/cgi-bin/SGD/getSeq?seq=YHR003C&flankl=0&flankr=0&map=a3map>
Genbank:
NC_001140- As for GPA1, the GenBank reference sequence information is contained within the sequence for chromosome 8 at this accession number.
NP_011866- The amino acid sequence for this hypothetical ORF (429 aa long) is located at this accession number.
SwissProt: <http://us.expasy.org/cgi-bin/niceprot.pl?P38756>
The information provided by SwissProt for the protein product of this gene indicates that the protein is 48.9 kDa, and shows strong similarity to yeast protein YKL027W. It also notes that there is some similarity between this protein and E. coli molybdenum cofactor biosynthesis protein A (MOAA).
BLAST Searches
The most significant result given by a search of NCBI BLASTn with the nucleotide sequence for YHR003C given at SGD is, as expected, the exact match from yeast chromosome 8. After this result, the remaining results are not very significant. There are two short (21-22 bp) alignments to mouse sequences about which no functional information is given.
A search of NCBI BLASTp with the protein sequence from accession number NP_011866 gives many significant results. Some of the most significant alignments are shown below:

Figure 4. BLASTp results for YHR003C. This list shows a selection of the most significant (E>100) results from the protein-protein BLAST for YHR003C. Image from: http://www.ncbi.nlm.nih.gov/blast/Blast.cgi Permission Pending.
From these results, it is evident that the most significant alignments are to other yeast proteins. YK1027Wp (accession number NP_012898) is also from Saccharomyces cerevisiae, but corresponds to a hypothetical ORF that has not been annotated. The next most significant alignment was to a protein (NP_593202) from Schizosaccharomyces pombe that is similar to the ubiquitin-activating enzyme of wheat. It is also noted that this protein has a Moeb/ThiF domain.
Many of the remaining alignments have ThiF domains or are involved in molybdopterin biosynthesis. It is likely that YHR003C has a role in yeast similar to the roles of these proteins in other organisms. Seeing the number of conserved domains suggests that a study of conserved domains might help to form a hypothesis of protein function.
Conserved Domains
A conserved domain search <http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=cdd> gives the following results for this protein:
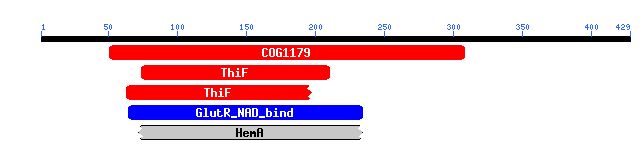
Figure 5. Conserved Domains in YHR003C. This image shows the conserved domains (colored boxes) found in the amino acid sequence of YHR003C (black line). Image from: http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi?RID=1065649181-19562-1029085.BLASTQ3 Permission Pending.
COG1179 is a domain seen in dinucleotide-utilizing enzymes that are involved in molybdopterin and thiamine biosynthesis.
The ThiF domain, which was also noted in the BLASTp results, is found in ubiquitin activating enzyme E1 and members of the bacterial ThiF/MoeB/HesA family.
GluR_NAD_bind indicates the presence of an NAD(P) binding domain similar to that from glutamyl-tRNAGlu reductase. Proteins with this domain use NADPH as a cofactor.
HemA, like the ThiF domain, is also seen in organisms that participate in coenzyme metabolism. The last two conserved domains (shown as blue and grey in the figure) are much less significant than the first two (shown in red)
Clusters of Orthologous Groups
Further investigation of the molybdopterin and thiamine biosynthesis conserved domain leads to the Clusters of Orthologous Groups (COG) <http://www.ncbi.nlm.nih.gov/COG/new/release/cow.cgi?cog=COG1179> The following dendrogram shows proteins that are closely related to YHR003C according to this conserved domain. As before, the other hypothetical yeast ORF (YKL027W) and the Schizosaccharomyces protein (SPAC1A6) are the most similar to YHR003C.
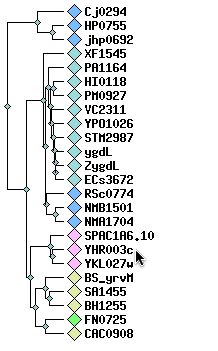
Figure 6. Cluster of YHR003C. This dendrogram shows proteins related to YHR003C (tip of cursor arrow) by its molybdopterin and thiamine biosynthesis conserved domain. Image from: http://www.ncbi.nlm.nih.gov/COG/new/release/cow.cgi?cog=COG1179 See also: http://www.ncbi.nih.gov/cgi-bin/COG/palox?seq=YHR003C and http://www.ncbi.nih.gov/cgi-bin/COG/blux?cog=YHR003c&YHR003c Permission Pending.
Kyte-Doolittle Predictions
A Kyte-Doolittle hydropathy plot of the amino acid sequence from NP_011866 gives the following results:
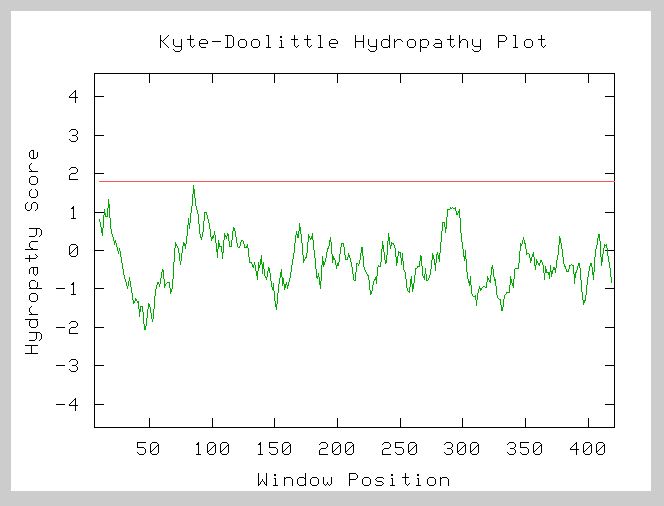
Figure 7. Kyte-Doolittle Hydropathy Plot for YHR003C. This graph is the hydropathy plot for the amino acid sequence of the hypothetical ORF. Points above zero indicate hydrophobic residues while points below zero represent hydrophilic residues. The window size is 19. The lack of strong peaks crossing the red line at 1.8 suggests that this protein is likely not an integral membrane protein. Image from: http://occawlonline.pearsoned.com/bookbind/pubbooks/bc_mcampbell_genomics_1/medialib/activities/kd/slrkd.pl Permission Obtained.
The lack of strong peaks above 1.8 in this plot indicates that YHR003C is likely not a transmembrane protein. The negative region from 25-100 predicts that this protein is globular and likely functions in the cytoplasm or within the hydrophilic region of some organelle of the yeast cell. This agrees with the information from the study of orthologous proteins. Proteins with the molybdopterin and thiamine biosynthesis conserved domain and the Thif domain are enzymes involved in biosynthesis and activation processes. These processes would not likely be carried out by integral membrane proteins. The initial peak may indicate an N-terminal signal sequence. This could mean that the protein is synthesized along the rough ER and then targeted to the interior of some specific cellular organelle.
Protein Associations
A search of the The GRID database of protein interactions for YHR003C gives the following information:
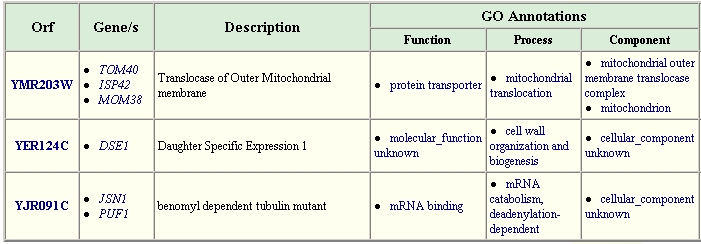
Figure 8. Proteins Interacting with YHR003C. This table lists proteins that were found to interact with YHR003C. Image from: http://biodata.mshri.on.ca/grid/servlet/SearchResults?keywords=YHR003C Permission Obtained.
Unfortunately, several aspects of the gene ontology for these associated genes is not known. However, what is known suggests that YHR003C might be a coenzyme (as supported by the conserved ThiF domain) for the processes of mRNA binding or mitochondrial protein transport. Since YER124C is involved in biogenesis, it makes sense that a protein like YHR003C that has conserved domains like those of proteins involved in also may be involved in molybdopterin and thiamine biosynthesis might aid in the biogenesis process.
Prediction of Function
From the information gathered, I would predict that YHR003C is a globular protein which functions as an enzyme. It may have catalytic activity involving nucleotide binding and be active in pathways involving biosynthesis.
References
Blackwell E. et. al. 2003. Effect of the pheromone-responsive G(alpha) and phosphatase proteins of Saccharomyces cerevisiae on the subcellular localization of the Fus3 mitogen-activated protein kinase. Mol Cell Biol. 23: 1135-50. <http://mcb.asm.org/cgi/content/full/23/4/1135?view=full&pmid=12556475> Accessed 2003 4 Oct.
Cherry JM. et. al. 1997. Genetic and physical maps of Saccharomyces cerevisiae. Nature 387: 67-73.
Dolinski, K. et. al. 2003. Saccharomyces Genome Database. <http://www.yeastgenome.org/> Accessed 2003 4 Oct.
Dumont, M. 2003. University of Rochester Medical Center Faculty Profile. <http://dbb.urmc.rochester.edu/bcbp/members/faculty/Dumont_Mark.HTMl> Accessed 2003 9 Oct.
Gene Ontology Software Group. 2003. Gene Ontology Database.<http://www.godatabase.org/dev/database/> Accessed 2003 5 Oct.
King, K. et. al. 1990. Control of yeast mating signal transduction by a mammalian beta 2-adrenergic receptor and Gs alpha subunit. Science 250: 121-3.
Li, E. et. al. 1998. Substitutions in the Pheromone-Responsive Gß Protein of Saccharomyces cerevisiae Confer a Defect in Recovery from Pheromone Treatment. Genetics 148: 947-961. <http://www.genetics.org/cgi/content/full/148/3/947> Accessed 2003 8 Oct.
Mount Sinai Hospital. 2002. The GRID database. <http://biodata.mshri.on.ca/grid/servlet/Index> Acccessed 2003 6 Oct.
NCBI. 2003. National Center for Biotechnology Information. <http://www.ncbi.nih.gov/> Accessed 2003 4 Oct.
SwissProt. 2003. Swiss-prot protein knowledgebase. <http://us.expasy.org/sprot/> Accessed 2003 5 Oct.
Yvert, G. et. al. 2003. Trans-acting regulatory variation in
Saccharomyces cerevisiae and the role of transcription factors. Nat. Genet.
35: 57-64.
Questions or Comments? Please e-mail Rachel Patton McCord
Rachel Patton McCord's Genomics Homepage