Image from Wikimedia commons
This website was produced as an assignment for an undergratuate course at Davidson College
Home safsaProtein dsafOrthologssdaf Jmol Structurefdas Review Paper

Image from Wikimedia commons
What Are iPS Cells and Why are They Important?
Embryonic stem (ES) cells have recently become the focus of intense research because they may have the potential to treat a multitude of diseases. ES cells are undifferentiated, which means they have the ability to become any other cell type in the body (they are pluripotent). The means that if they could be induced to become liver cells they could treat someone with hepatitis, but they could also become skin cells and help burn victims. Until the ES cells differentiate, they can grow indefinitely in culture. Normally once a cell differentiates, for example into a red blood cell, it will always be a red blood cell; there is no regressing. In their paper, "Induction of Pluripotent Stem Cells from Mouse Embryonic and Adult Fibroblast Cultures by Defined Factors," Takahashi and Yamanaka present evidence to suggest that if the right factors are introduced cells revert back to a pluripotent state. They called these reverted cells, iPS cells. If iPS cells really are like ES cells, they could be used for therapeutic treatment without causing the controversy that ES cells do.
How Did They Create iPS Cells?
Takahashi and Yamanaka chose twenty-four candidate genes to test for the ability to induce pluripotency. They selected these genes based on the fact that most have previously been shown to be important in ES cell life. They wanted to be able to easily tell if pluripotency had been achieved, so they inserted a combination β-galactosidas and neomycin resistance gene behind the Fbx15 promoter (Fig. 1a). The Fbx15 gene is only expressed in ES cells and embryos, so if the cells are resistant to neomycin, they must be expressing Fbx15 and therefore be similar to ES cells. When they transducted all twenty-four candidate genes into mouse embryonic fibroblasts (MEFs) they did get some colonies that were resistant to the neomycin (Fig. 1b). A closer look at the colonies, called induced pluripotent cells (iPS) shows that they look very similar to ES cells, and nothing like the MEF cells they used to be (Fig. 1c). The doubling time of the iPS cells is also on par with ES cells, not MEF cells (Fig. 1d). All in all this portion of the figure (Fig. 1b-d) is very convincing. The cells are alive, they look right, and they’re dividing rapidly.
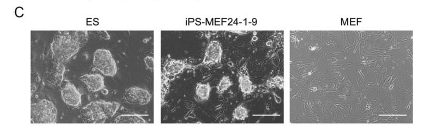
Figure 1c: A comparison of the morphology of ES, iPS, and MEF cells. Notice that the iPS cells look like the ES cells even though they used to be MEF cells.
The remainder of the figure (Fig. 1e-f) should be the focus of more research in the future. Takahashi and Yamanaka used RT-PCR to look at the expression levels of different genes that are typically expressed in ES cells (Fig. 1e). They compared the levels in three different iPS colonies with that of ES and MEF cells. It is true that the iPS cells more closely resemble ES cells, but just given this figure I would not be convinced. The levels of Nanog, Fgf4, Cripto, and Zfp296, are very similar between the ES and iPS cells. However Eras, Oct3/4, Sox, and Dax1, all appear significantly different. Then they looked at methylation of the genes themselves (Fig. 1f). They found that Oct3/4 was heavily methylated in the iPS cells, as in MEF cells, whereas it is not methylated in ES cells. Nanog and Fbx15 were unmethylated in iPS and ES cells, but methylated in MEF cells. These results do make sense when taken with the results of the RT-PCR; methylation of the gene tends to inhibit transcription. However I think it would have been beneficial to have examined the methylation of the other genes that were not as strongly expressed in the iPS cells (so Eras, Sox2, and Dax1).
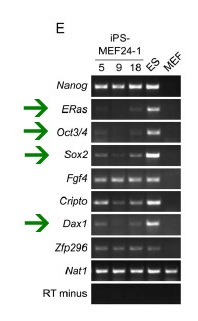
Figure 1e: The results of an RT-PCR of ES cell marker genes. The green arrows highlight genes that do not show the same expression in iPS and ES cells. Nat1 is a loading control and RT minus is a negative control.
After determining that some combination of the twenty-four candidate genes induced these cells to revert to their pluripotent form, Takahashi and Yamanaka wanted to narrow down the factors to only the crucial ones. Therefore they looked to see what genes, if they were removed, would inhibit the formation of colonies. They found ten different factors that led to very little or no colony growth (Fig. 2a). The importance of those ten factors was further emphasized by the fact that if only they were transducted into MEF cells more colonies formed than when all twenty-four were transducted (Fig. 2b). They performed the same removal of each factor individually from the ten and ultimately came up with the four most important factors (Oct3/4, Sox2, c-Myc, and Klf4). Further attempts to narrow it down to combinations of two or three factors were relatively unsuccessful (Fig. 2c). They did find that without Sox2 cell colonies formed and could be maintained, however they do not look like ES cells (Fig. 2d). They say that without c-Myc colonies that do not look like ES cells form, so I found it curious that they only continued their investigation on the –Sox2 colonies and not the – c-Myc colonies. They do not explain why they chose the -Sox2 over the -c-Myc.
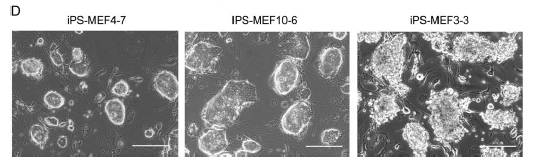
Figure 2d: Morphological comparison of different iPS cell colonies. Notice that the two on the left (MEF4 and MEF10) have cells with a smooth, round shape. The cells on the right (the -Sox2 colonies) have jagged uneven edges.
Once again Takahasi and Yamanaka used RT-PCR to look at ES marker gene expression in the iPS cells. They compared iPS-MEF3 (-Sox2), iPS-MEF4 (Oct3/4, Sox2, c-Myc, and Klf4), and iPS-MEF10 (the 10 factors) with ES and normal MEF cells (Fig. 3a). Like in their first RT-PCR, they found that overall iPS cells resemble ES cells, but they are not identical. They chose two clones that most closely resembled ES cells and examined them further. First they looked at the methylation and acetylation of the Oct3/4 and Nanog promoters (Fig. 3b). They found that the promoters for these genes remained partially methylated in the iPS cells (Fig 3c). Differential methylation and acetylation can result in differential expression, so the mechanisms behind why some genes are de-methylated and others are not is definitely an area for future research. Takahashi and Yamanaka are very honest and say that these results show that the iPS and ES cells are not identical. In addition to the ES marker genes, they went on to look at global gene expression using DNA microarrays. Using statistical analysis (Pearson correlation), they found that, in terms of overall gene expression, iPS cells are closer to ES cells than MEF cells (Fig. 4). The microarrays also further confirmed their suspicions (from Fig. 3a), that iPS-MEF3 cells are significantly different than iPS-MEF4 or iPS-MEF10 cells.
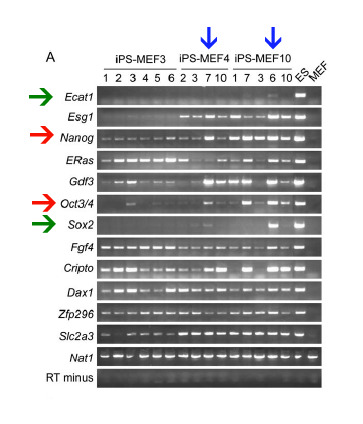
Figure 3a: RT-PCR results comparing iPS cells to ES and MEF cells. Again the genes being looked at are ES cell marker genes. The green arrows point out the two genes that though strongly expressed in ES cells, are barely expressed in iPS cells. The red arrows point to the two genes that Takahashi and Yamanaka further investigated. The blue arrows point to the two iPS cell lines that they thought most closely resembled ES cells.
At this point Takahashi and Yamanaka have shown that iPS cells generally express ES cell markers, but they have yet to show than iPS cells are pluripotent. They do so by injecting the cells into mice. Where they injected the cells, teratomas formed (Fig 5a). A teratoma is a tumor than contains cells from all three germ layers (Fig. 5b). In addition to forming teratomas, the iPS cells were able to form embryoid bodies (Fig. 5c). Embryoid bodies are aggregates of cells that form from stem cells. Only iPS-MEF4 and iPSMEF10 embryoid bodies differentiated into all three layers (Fig. 5d).
Once they had determined the iPS cells are pluripotent, Takahashi and Yamanaka looked to see if the iPS cells, when introduced to a blastocyst could become part of the growing mouse (as a normal ES cell could). To do this they took tail tip fibroblast cells from a mouse that was expressing green fluorescent protein (GFP) and introduced Oct3/4, Sox2, c-Myc, and Klf4. The colonies they obtained looked the same as ES cells (Fig. 6a). They looked at ES gene marker expression in the iPS-TTFgfp4 cells using RT-PCR, as they had previously done (Fig. 6b), as usual the iPS cells resemble but are not identical to ES cells. They injected some of the iPS-TTFgfp4 cells into a normal mouse blastocysts and actually observed normal embryo development (Fig. 6c). Looking at all three germ layers, they found cells in each expressing GFP (Fig. 6d). This seems to suggest that iPS cells can contribute to development just as normal ES cells do.
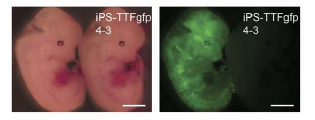
Figure 6c: The development of blastocysts injected with iPS-TTFgfp4 cells into embryos. On the right you can see the flourescence throughout the embryo, indicating that cells expressing GFP (iPS cells) have been incorporated.
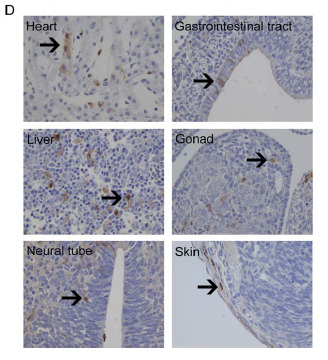
Figure 6d: Immunostaining for GFP in all three germ layers. The arrows point to brown staining, which indicates the presence of GFP.
Takahashi and Yamanaka were concerned that introducing the four factors, and therefore increasing the gene amount, in iPS cells might cause an over expression of the gene products. However when they did a Western blot, they found that the total protein in ES and iPS cells was almost the same (Fig 7a). The levels of Nanog and E-Ras were lower in iPS cells than in ES cells, which demands further investigation. They also looked at where a particular gene (Klf4) had integrated into the genome (Fig. 7c). They found that the integration was in a different location each time.
Critiques of the Paper?
Parts of figure 7 (Fig. 7c-e) are characteristic of the paper as a whole. Takahashi and Yamanaka wanted to exclude the possibility that somehow what they thought were iPS cells were actually just ES cells that had contaminated their sample. In other figures there were usually good positive and negative controls, along with examples of both ES and MEF cells for comparison. In Fig. 5 I would have liked to see comparable results from ES cells, especially as I have no idea what any of the pictures should actually look like. Overall I think given the results they have Takahashi and Yamanaka make the right sort of claims. Calling them iPS cells instantly distinguishes them from ES cells, which is what should be done. Multiple times throughout the paper they mention that although the cells are similar, they are not identical. They also admit that right now they essentially have no idea why these four factors are sufficient, or how the reversion to pluripotency actually occurs. The biggest critique I have of the paper is I think they downplay the differences in expression of ES marker genes. They do show (Fig. 7a) that the levels of protein of the introduced genes are essentially equivalent between iPS and ES cells. However given the results in Fig. 3a and Fig. 4, I think it is clear there are significant differences in the composition of proteins produced by each cell type.
What's Next?
As mentioned in the introduction much of the interest in ES cells has surrounded the potential for therapeutic treatment. The potential to induce pluripotency in human cells is obviously a desirable goal. If people's own cells could become iPS cells they could be used without fear of rejection. Therefore a clear next area of research is an attempt to reproduce the results Takahashi and Yamanaka got with mouse cells in human cells. In 2007 the same lab did just that (Takahashi et al., 2007). They found that using that same four factors they used in mice, they could generate iPS cells from human fibroblasts.
Interestingly at almost the same time Yu et al. (2007) also generated iPS cells from human somatic cells, however they used a different set of four factors (Oct4, Sox2, Nanog, and Lin28). Obviously neither group understands exactly what effect the expression of each gene is causing in the cell. Therefore I think future experiments might step back slightly from the creation of iPS cells and focus on the individual function of each protein product. This could be done by transducting each gene individually and not selecting for pluripotency, but rather just observing the effects of the protein. This could include things such as localization, methylation of ES marker genes, and global gene expression. In this way we might be able to narrow down exactly what needs to happen for pluripotency to be restored. Before the iPS cells are clinically applicable, I think a lot more research needs to go into epigenetic regulation. As Takahashi and Yamanaka saw in Figure 1f when the four factors were introduced some genes were demethylated while others were not. I would use the same bisulfite sequencing technique they used to look at the methylation status of all the ES marker genes that were differentially regulated in iPS cells. I suspect that most of those genes would show the same sort of pattern that Takahashi and Yamanaka found for Oct3/4.
Currently a drug, Vidaza (azacitidine), is approved by the FDA for the treatment of myelodysplastic syndrome. When used in vitro Vidaza removes methyl groups from DNA. Vidaza is an analogue of cytosine, which is a component of RNA and DNA. When Vidaza is present in cells during replication it is incorporated into the DNA instead of cytosine, and then the DNA cannot be methylated. Methylation typically occurs later in life as part of epigenetic regulation, so perhaps incorporating Vidaza with the four factors could create cells that have methylation profiles more similar to ES cells. I would create iPS cells just as Takahashi and Yamanaka did, only with doses of Vidaza present during the process. Then I would compare the methylation status of ES marker genes in the Vidaza iPS cells with the methylation status determined in the prior suggested experiment.
Email me: capiper@davidson.edu