MacDNAsis was used to analyze the
amino acid and cDNA sequences for MAP kinase in five organisms. These
sequences were obtained from a Genbank search and can be accessed below.
Homo
sapiens
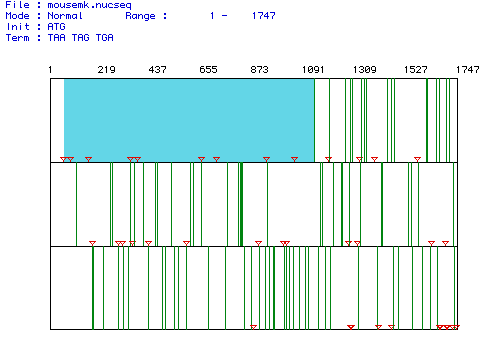
The largest open reading frame (ORF) for the cDNA encoding Mus musculus MAP kinase includes nucleotides 61-1137. In the figure below red triangles represent start codons, green lines represent stop codons, and the blue highlighted region is the largest ORF.

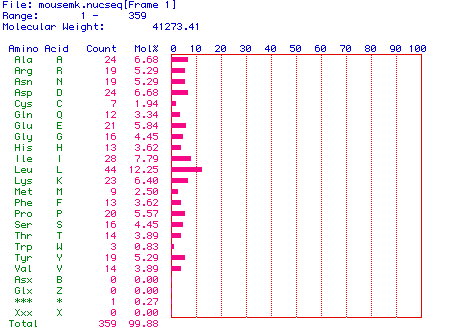
When MacDNAsis was used to translate the largest ORF (61-1137),
a predicted molecular weight of 41273.41 was
yielded (see below).
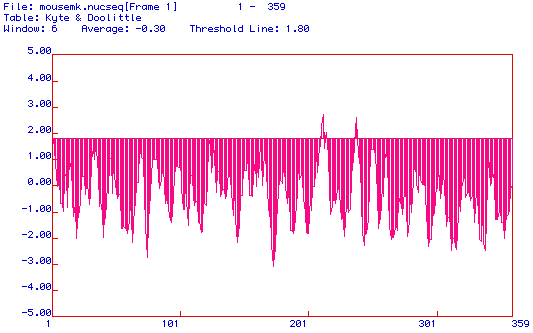
Below is a Kyte-Doolittle hydropathy plot of MAP kinase.
Positive values indicate hydrophobic regions, while negative values are
indicative of hydrophilic regions. A horizontal threshold line has been
inserted at 1.8 to reveal the predicted transmembrane domains. The
generally accepted value used to determine the presence of transmembrane
domains is 2. It appears therefore that MAP kinase has 2 probable
transmembrane domains and as a result is likely to be an integral protein.
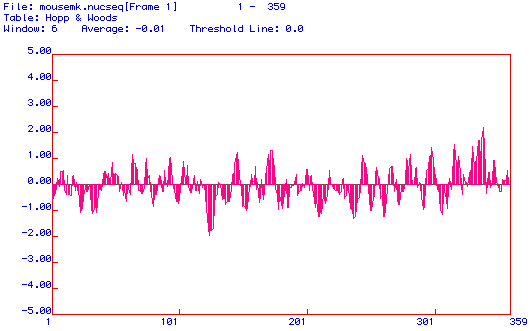
Below is an antigenicity plot (also called a Hopp and Woods plot) which indicates the portions of the protein that would be used to generate a peptide if one wanted to make a monoclonal antibody against the peptide. The portiion of the protein that would be used to generate a peptide would be near amino acid 345 since this represents the most hydrophilic region of the protein.
To see the predicted secondary struture of MAP kinase
click
here.
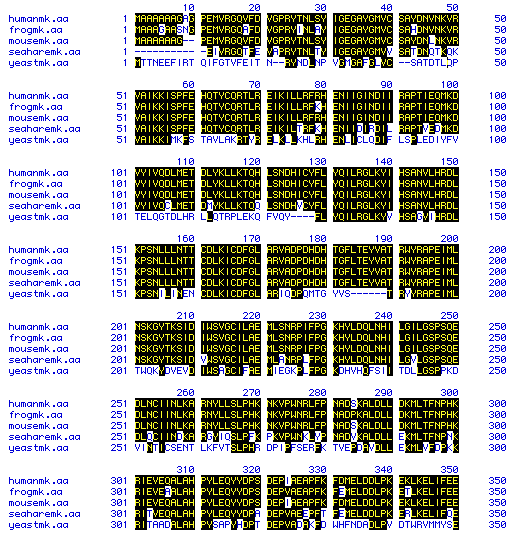
MacDNAsis was also used to perform a multiple sequence alignment of MAP kinase for all five organisms examined. As one would expect, the greatest homology exists between the mammalian representatives Homo sapiens and Mus musculus. However, the sequences of the frog Xenopus laevis and the sea slug Aplysia californica also possessed a high degreee of sequence similarity. The sequence from Saccharomyces cerevisiae however, is clearly divergent from the other organisms. It should be mentioned that the amino acid sequence from Aplysia californica is a partial coding sequence, which may be a possible explanation for its divergence.
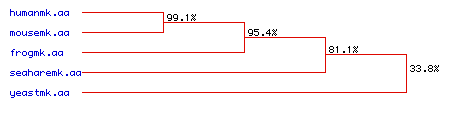
Below is a phylogenetic tree which depicts the degree
of amino acid conservation over time. Again we see that there is
a high degree of sequence similarity between between Homo sapiens,
Mus
musculus, and Xenopus laevis. The amino acid sequence
encoding MAP kinase in Aplysia californica is
also relatively conserved.
However, the amino acid sequence of the yeast
Saccharomyces cerevisiae is far more divergent
Click on one of the organisms below to view the amino acid and cDNA sequences encoding MAP kinase.
Homo
sapiens
Mus
musculus
Xenopus
laevis
Aplysia
californica
Saccharomyces
cerevisiae