Methods Used by GCAT
Members
General Resources
GCAT
Workshop-Produced, Student-Tested Protocols
The "Indirect Dye"method uses
amino allyl dUTP to label the cDNA. This method prevents the dyes
from creating a bias in the cDNA synthesis. The "Direct Dye"
method uses Cy3/Cy5-coupled dUTP for cDNA synthesis. This reduces
the number of steps to make the labeled cDNA. The 3DNA method uses
Genisphere's dendrimer with about 200 dyes per cDNA so the signal
is very strong. You can download the methods we used, and personal
comments from the table below:
Tips we learned that
help for all three microarray methods
|
Validate your candidate genes with Quantitative PCR
without Real-Time PCR. (Word File or PDF
File)
This is a time line and protocol that you can use. It goes
with the paper
by Bradford et al. 2005.
Watch a
short movie (~ 5 min) to see how the microarrays
are scanned here at Davidson.
Main
Brown Lab Protocol Page
Direct Labeling of RNA with
Alexa Dyes (Molecular
Probes Kit): Protocol from the Institute for Systems Biology in pdf format
Hybridization and washing
conditions for Alexa Method (see above): Protocol from the Institute
for Systems Biology in pdf format.
Modified Protocol
for creating cDNA probes with Alexa Dyes, hybing and washing. Collaborative
effort with GCAT member Todd Eckdahl and Alison Golden at the Institute for
Systems Biology
Online
Dry-Lab to Explore DeRisi Experiment Teaching resources for working
through the DeRisi diauxic shift paper (1997) using the original data. Ideal
for allowing students to compare their analysis with the published version.
All components are free and can be used with MAGIC
Tool, free microarray software.
Trouble-shooting tiff files (from 2009
workshop)
Lowering your coverslip to maximize mixing of hybridization
buffer (contributed by Cynthia Horst, Carroll College)
SOP for MAGIC
Tool (sequential set of steps for typical situations)
Software Protocols
Scanalyze Protocol and
Advice
GeneSpring
Online Tutorial (produced by Silicon
Genetics)
GeneSpring
Protocol. Take your data from Scanalyse to GeneSpring. Good for looking
at a series of related experiments (e.g. time course, dose response, etc.)
This protocol was developed by Terrie Rife at James Maddison University.
Posting GCAT Datasets
on SMD (Stanford Microarray Database)
Accessing the ISB FTP Server
Clustering Protocol
Free Download
of PC Software for Analysis of Scanned Chip
(from Mike Eisen's site; login required)
MAGIC Tool
- free software, works on all computer platforms and goes from tiff
files to clustering
Pedagogical Resources
Time
line for teaching with DNA microarrays (excel file).
This 6 week time line was developed by 4 seasoned GCAT memebrs: Lauire Caslake,
Lafayette College <caslakel@mail.lafayette.edu>;
Myra Derbyshire, Mount Sant Mary's College <derbyshi@msmary.edu>; Jeff
Newman, Lycoming College <newman@lycoming.edu>;
Amy Vollmer, Swarthmore College <avollme1@swarthmore.edu>.
Educational
Site for Online Clustering (Interactive
with real-time clustering)
Online
Dry-Lab to Explore DeRisi Experiment Teaching resources for working
through the DeRisi diauxic shift paper (1997) using the original data. Ideal
for allowing students to compare their analysis with the published version.
All components are free and can be used with MAGIC
Tool, free microarray software.
The
The Polyploidy Portal has many resources related to multiple
copies of chromosomes. In addition, this multi-campus group has produced
a hands-on
Excel-based module to help students learn about microarray data analysis.
These are all freely available.
Complete
(Soup to Nuts) description of how to use yeast for DNA microarray experiments.
Intended for the novice teacher, Dave Kushner's 20 page protocol offers
advise and wisdom. You can use his method for RNA isolation, or others
provided by GCAT members
(method 1, method
2, QC of RNA)
Yeast Resources
Download Gene List for spring 2006/2007
through 2011/2012 Yeast Chips produced from Washington Univ.
(St. Louis); 70mer Illumina oligos
printed on epoxy slides.
Download Gene List for fall 2006/2007 shipment
of Yeast Chips produced at Washington Univ. (St. Louis); 70mer oligos printed
on epoxy slides.
Download Gene File for 2005/2006 Yeast Chips produced
at Washington Univ. (St. Louis); 70mer oligos printed on epoxy slides.
.
Download Gene File for 2004/2005 Yeast Chips produced
at Washington Univ. (St. Louis); 70mer oligos printed on epoxy slides.
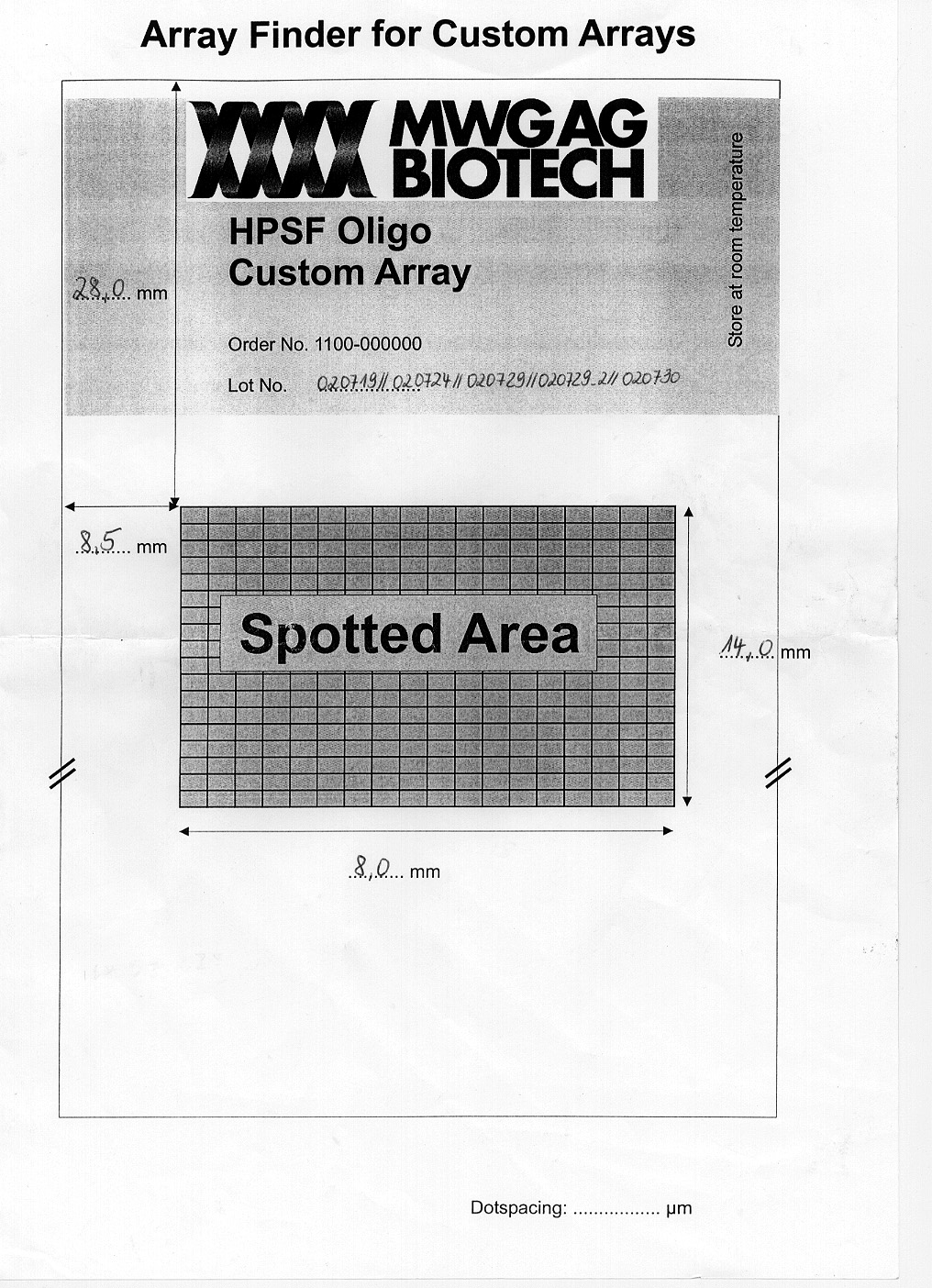
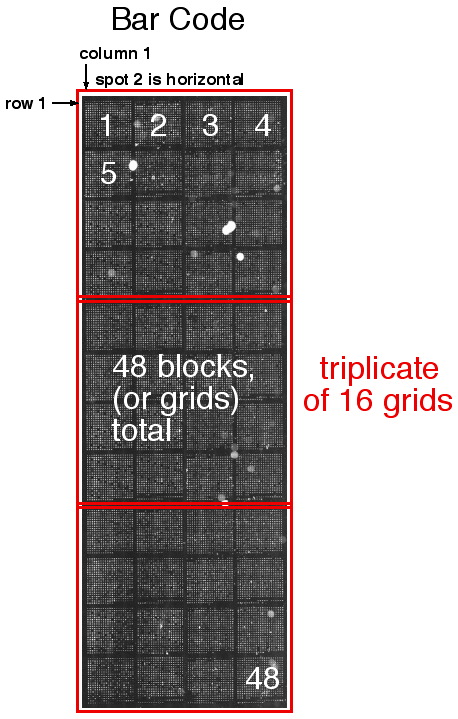
How to orient the spots:
Turn the slide so that it is a tall rectangle with the slide
number etched at the bottom of the slide, facing up. Then, the Top half has 16 grids
(with the Bottom half being a duplicate). Then, the grids are as follows:
1 |
2 |
3 |
4 |
5 |
6 |
7 |
8 |
9 |
10 |
11 |
12 |
13 |
14 |
15 |
16 |
1 |
2 |
3 |
4 |
5 |
6 |
7 |
8 |
9 |
10 |
11 |
12 |
13 |
14 |
15 |
16 |
|
Slide Number
(DNA side Up)
|
Within each grid, the 2nd spot is to the right of the 1st spot,
which is in the top left of each grid.
Basically, once you have the slide oriented, everything is just like a
Book...
Download Gene File for Stanford Y01 and Y02 series of chips
Download Gene Files for these ISB chips
MWG HPSF Custom Array A 78 series
Making Media and Growing
Yeast
Isolating Yeast RNA /
mRNA
3DNA Method for Making
Probes, Pre-Hybe, Hybe, Wash for Microarray Hybridization
(developed by GCAT
and Genisphere, NJ)
Hot Phenol RNA isolation and
3DNA Method from beginning to end (very good results) - David
Kushner at Dickinson College
PDF Version (4 MB) or
RTF version (16 MB) or
Word File version (1
MB)
Griding on ISB Chips
ISB Oligo-Array Information
(Word file)
Yeast Bar Coded Resources
- no longer available
You can read a bit about the bar
code method at this web page. We have two pools of mutant cells: heterozygotes and homozygotes.
The homozygotes have both alleles deleted and there are about 4200 strains
pooled into one batch of cells. The heterozygotes have one allele deleted
by the bar code method, and the other allele is wt. These are heterozygotes
because homozygote deletions are lethal; we have about 1800 heterozygote
strains pooled in one batch of cells. We have frozen stocks of these and
can send you what we have as long as they last. We obtained these as a
generous donation from Corey
Nislow who is at the Univeristy of Toronto. The bar code microarrays
are provided directly from Agilent to the end user. GCAT is
billed for these chips and GCAT bills
the end user our normal costs. The list of strains and PCR primers used
to amplify the bar codes from genomic DNA are in the Excel files below.
Arabidopsis
| Gene Files since 2007 Arabidopsis Chips
produced at University of Arizona. |
|
Download
gene file starting August 2009 (batch AT3.8.2.Z; tab delimted text
format = MAGIC Tool ready)
Download gene
file starting August 2009 (batch AT3.8.2.Z;
Excel File.xls)
Download gene
file starting August 2009 (batch AT3.8.2.Z;
Gal file.gal)
Download
Gene file starting January 2009 (tab delimited text
format = MAGIC Tool ready)
Download Gene file starting January 2009 (Gal
format)
Download
Gene file starting January 2009 (excel
format .xlsx)
Download Gene
File for 2007-2008 (tab delimited text format = MAGIC Tool ready)
Download Gene
File for 2007-2008 (Gal format)
Download Gene
File for 2007-2008 (excel format)
List of genes,
some chromosomal information, and some GO information. NOT ready for
MAGIC Tool yet. Contributed by Bruce Kohorn at Bowdoin College.
Information about all Arabidopsis microarrays
since 2003
Gene File for 2006/2007 Arabidopsis Chips produced
at University of Arizona. |
 |
Download Gene
File for 2006-2007 (tab delimited text format = MAGIC Tool ready)
Download
Gene File for 2006-2007 (Gal format)
Download Gene
File for 2006-2007 (excel format)
Information about all Arabidopsis microarrays
since 2003
Download Gene File for 2005/2006 Arabidopsis Chips produced
at University of Arizona
Information about all Arabidopsis microarrays since 2003
at http://www.ag.arizona.edu/microarray/
Download Gene File
for fall 2001
Chicken (new 2007)
Download Gene File for 2007/2008 Chicken Chips produced
at University of Arizona
Download Gene
File for 2007-2008 (tab delimited text format = MAGIC Tool ready)
Download Gene
File for 2007-2008 (Gal format)
Download Gene
File for 2007-2008 (excel format)
Information about all
chicken microarrays
Drosophila
2011-2012 Fly chips were produced at UHN Microarray Centre. You need these files:
2009-2011 Fly chips were produced at the Canadian Drosophila Microarray
Centre. You can download
all the 14kv1 files you need at this URL.
Layout
for the spot pattern. (PDF file)
2007/2008 Fly Chips produced at University
of Oregon have NO LABELS! Please fog them and put a label (etched) on the
same side as the DNA. Then when you send the slides for scanning, be sure
to tell us where the label is relative to DNA. We will have to figure out
the orientation as we go along.
An email said "Jason sent the arrays, so I'm not
sure what he did in the way of etching. Tell them that if they look
at an array and each block looks like this:
..................
..................
..................
.........
"
|
Campbell suggests:
I strongly suggest you
fog the slides and look at them to try to match this pattern with genelist.
Then you will know where to put your etch mark so we can scan
them in the normal orientation. However, keep in mind that MAGIC
Tool allows you to indicate the correct order for numbering,
so you can recover from any orientation mistakes if you are careful
at the time of addressing
the slide. |
They are NOT post-processed
yet. Download post-print
processing protocol. This is different from last year's chips.
Download Gene File
for 2007-2008 (tab delimited text format = MAGIC Tool ready)
Download Gene File
for 2007-2008 (Gal format)
Download Gene File
for 2007-2008 (excel format)
Download Gene File for 2006/2007 Fly Chips produced at University
of Oregon
They are long, amino-modified oligo arrays on
aldehyde slides; oligos synthesized by Illumina for
a consortium of fly lab called INDAC.
- Chips are from the beginning of the print run, so some spots
might be a little "blobby",
but overall they are very nice arrays.
- I etched the back of the array and numbered them. The back does NOT have
the DNA.
- They are NOT post-processed yet. Download post-print
processing protocol.
Download Gene File
for 2006-2007 (tab delimited text format = MAGIC Tool ready)
Download Gene File
for 2006-2007 (Gal format)
Download Gene File
for 2006-2007 (excel format)
Download Gene File for 2005/2006 Fly Chips produced
at University of Oregon
They are long oligo arrays, synthesized by Illumina for a consortium
of fly lab called INDAC.
The DNA is up, the spots are read from left to right. The info in the
GAL file give the predicted fly gene "CG####" for each spot. If
everything is lined up right, the first row of each block should be mostly
control spots that give little signal (Arabidopsis DNA), and have a non-CG####
identifier.
- The lab that made these slides uses 25x60mm "lifterslip" coverslips
(available through Erie Scientific, Fisher and
VWR).
- They use 40 μL hyb solution per array.
- The slides are poly-L-lysine (he thinks).
- The oligos are 65- to 70-mers.
- The arrays have been post-processed, so steaming and snap-drying shouldn't
be necessary. You can verify this by looking for spots. If you do not see
them, but you can see them when you fog the slide with your breath, then
the chips have been post-(print)-processed.
- They're better "fresh" (i.e. used soon) but they are probably
good for "a while".
- They should tolerate higher hyb temps (55-65 degrees) but they've only
used formamide buffer and 42 degrees themselves.
Download Gene File
for 2005-2006 (tab delimited text format)
Download Gene File
for 2005-2006 (Gal format)
Download Gene File
for 2005-2006 (excel format)
Download Gene File for 2004/2005 Fly Chips produced at
University of Oregon
Download Gene File
for 2004-2005 (tab delimited text format)
Download Gene File
for 2004-2005 (Gal format)
Download Gene File
for 2004-2005 (excel format)
Download Gene File for
2003-2004 (excel format)
Download Gene File for
2003-2004 (tab delimited text format)
E. coli
E.
coli chips for 2006 - 2011 were produced at the
University of Alberta.
UW-Madison no longer produces microarrays.
Download Gene File for 2009-2011 E.
coli Chips produced at University of Alberta
Cover
Letter explaining everything (Word File)
How
to work with epoxide slides to reduce background
MAGIC
Tool ready gene list and Excel
version and .GAL version
Download Gene File for 2008/2009 E.
coli Chips produced at University of Alberta
Cover Letter explaining everything
Compressed CD contents except images (.gal file,
orientation on chips, processing of slides, etc.)
Compressed CD images
MAGIC
Tool ready gene list and Excel version
Download Gene File for 2007/2008 E.
coli Chips produced at University of Alberta
E.
coli gene
list and layout, (MAGIC Tool ready, tab delimted text file, "b
numbers" in first column)
E.
coli gene
list and layout, (.gal format for GenePix)
E.
coli gene
list and layout, (Excel format for orientation)
File
about Slide Preparation - NO UV X-linking Needed (General_Notes_Epoxide.doc)
List of Oligos and annotation (ecoli_oligos_input.xls)
Quality Control slide (9mer hybed: Panomer_9_w595.tif)
Download Gene File for 2006/2007 E.
coli Chips produced at University of Alberta
E.
coli gene
list and layout, (MAGIC Tool ready, tab delimted text file, "b numbers" in
first column)
E. coli gene list and
layout, (.gal format for GenePix)
E. coli gene list and
layout, (Excel format for orientation)
Additional files for 2006/2007 E.
coli Chips produced at University of Alberta
E.
coli Word
file for overview of these chips
(Including
pre-hybridization protocol to reduce background)
PDF
file about the type of slides used (Epoxide)
Slides DO NOT need to be cross-linked by UV light or baking.
List of all gene names (synonyms), oligo sequences, functional
information
Download Gene File for 2005/2006 E.
coli Chips produced at University of Wisconsin - Madison
E. coli (ECO19)
product data sheet (word file format)
E. coli (ECO19)
gene list and layout, 4 sheets (excel file format)
E. coli (ECO19)
gene list and layout, 1 sheet (MAGIC Tool ready, tab delimted text file, "b numbers" in first
column)
Download Gene File for 2004/2005E.
coli Chips produced at University of Wisconsin - Madison
Download
Gene File for Fall 2004 (ECO18 series) excel file, 3 sheets
Download
Gene File for Fall 2003 (ECO17 series) excel file, two sheets
Download
Gene File for Fall 2003 (ECO16 series) excel file
Download
Gene File for Fall 2002 (ECO15 series) excel file
Description
of E. coli chips (fall 2002) Word File
Download
Gene File for Fall 2002 (ECO14 series) excel file
Download Gene File
for fall 2001
E.
coli Hyb Setup.pdf
E. coli
genomic DNA labeling.pdf
E. coli
RNA isolation.pdf
E.
coli RNA
labeling.pdf
E.
coli indirect
labeling (BIPL).pdf
E.
coli
indirect labeling with EFA.pdf
Reprint of paper by Don Court et al. "An
efficient recombination system for chromosome engineering in Escherichia
coli" PNAS Vol. 97 (11): 5978-5983. May 2000. In Adobe Acrobat
Format
Protocol
for performing the method descibed in paper above by Don Court. In
Word format.
Human
Download Gene File for
2010-2011 Human chips produced at Phalanx
Download gene
file starting fall 2010 (excel format)
Download gene
file starting fall 2010 (MAGIC Tool ready, tab delimited text format)
Download
gene file starting fall 2010 (gal format)
Additional gene information available at Phalanx
Download Gene File for
2007-2010 Human HEEBO Chips produced at Washington University,
St. Louis
Using MAGIC Tool on human DNA microarrays (Word File)
(student instructions written by Michael Hamann, Assistant Professor of Biology, Bemidji State University, MN)
Download gene
file starting fall 2007 (excel format)
Download gene
file starting fall 2007 (MAGIC Tool ready, tab delimited text format)
Download
gene file starting fall 2007 (gal format)
Here is the conversion file (HEEBO_Human_Set_v1.00)
for identifying the genes from the oligo names (compare columns F and S).
Also, sequecnes for the oligos are available here. This file is in Excel
format of .xlsx. The
file is very large, so it will take a while.
Download Gene File for
2006/2007 Human HEEBO Chips produced at Washington
University, St. Louis
Download gene
file starting fall 2006 (excel format)
Download gene
file starting fall 2006 (MAGIC Tool ready, tab delimited text format)
Download gene
file starting fall 2006 (gal format)
Download Gene File
for 2005/2006 Human HEEBO Chips produced at Washington
University, St. Louis
Download gene
file starting fall 2005 (excel format)
Download gene
file starting fall 2005 (tab delimited text
format)
Download gene
file starting fall 2005 (gal format)
From Daron Barnard at College of the Holy Cross: Gene
List in Gal Format (3 MB .gal file that opens in Excel if needed)
On the gene list file there
are two columns with identifiers: one is a oligo ID that is not helpful in
searching a database, and then there is the 'Name' column. Some of these
entries have the GI # and the Accession # but they are buried in a FASTA
type format (without the <).
The program that I am using for the analysis of the files is CARMAweb (a
Web based GUI for analysis using R and the BioConductor packages) and after
clustering I can examine any specific gene's expression and link directly
to the gene page for that gene - but the gene list doesn't provide information
in a way that is easily accessible. Also I can do GO-analysis, but again
I need a column that has an identifier that the program recognizes.
In a nut
shell: this extracts the accession number for (many of the) genes (I am
not sure why some do not have accession numbers) in a way that can be easily
read by programs since they are in a separate column.
Download Gene File
for 2002 - 2004/2005Human Chips produced at University
of Miami Medical School
Download gene file
for 2002-2003, 2003-2004, and 2004-2005 (excel format)
Download gene file
for 2002-2003, 2003-2004 and 2004-2005 (tab delimited text format)
Download gene file
for 2002-2003, 2003-2004 and 2004-2005 (gal format)
Download Gene File for
fall 2001
Maize
Download
Gene File for 2007/2008 and 2008-09
Microarray Storage and Hybridization Preparation
- Make sure that
you have received the right slides that you requested.
- Store the microarrays
at room temperature protected from light in a dust free environment. Do not
store arrays in the cold room or above 50 °C.
- Long-oligo arrays can be
stored up to 12 months at room temperature. Do not forget the re-hydration
and UV crosslinking steps that are needed prior to hybridization.
- The DNA
oligos are printed on the labeled side of the slide. When exposing the
slides to either the water vapor for the re-hydration or UV light for crosslinking,
be sure to expose the DNA side of the slide.
- At no time before the crosslinking
step are the slides to be immersed in water. Slides may be stored for longer
periods of time after re-hydration and UV crosslinking.
Gene
List for 2007 - 2008 microarrays (text file, MAGIC Tool ready)
Gene
List for 2007 - 2008 microarrays (Excel file for exploring)
Gene
List for 2007 - 2008 microarrays (.gal file for GenePix)
Maize
Chips produced Iowa State University.
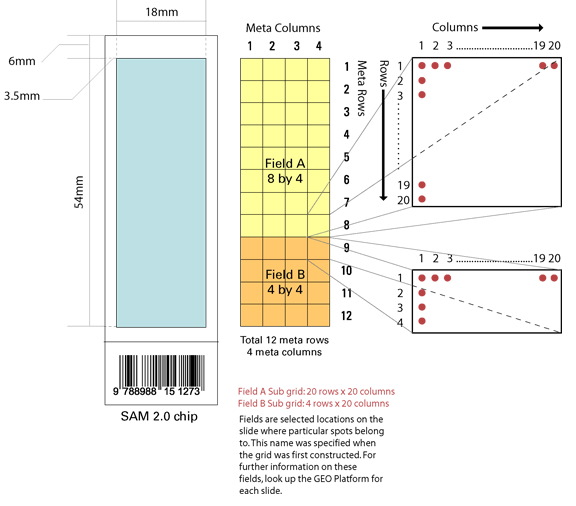
Download
Gene File for 2006/2007
Gene
List for 2006 - 2007 microarrays (text
file, MAGIC Tool ready)
Gene
List for 2006 - 2007 microarrays (Excel
file for exploring)
Gene List
for 2006 - 2007 microarrays (.gal
file for GenePix)
Description
of SAM2.0 Maize microarrays (PDF
file)
Download Gene
File for 2005/2006 Maize
Chips produced Iowa State University. A
range .gal files are available at this web site.
Gene List for
2005 - 2006 microarrays (text file, not ready for MAGIC Tool)
Gene List for
2005 - 2006 microarrays (text file, MAGIC Tool ready)
Layout
for microarray design 2005 - 2006 (PowerPoint Slide)
Additional files for 2006/2007 Maize Chips
produced at Iowa State University
Description of slides used for spotting (PDF)
Rat
Download Gene File for 2010-2011 Rat Chips produced at
Washington Univ. (St. Louis); oligos printed on epoxy slides.
Detailed description of rat genes and code names used for oligos
Mouse
Download Gene File for 2007 - 2011 Mouse Chips produced at
Washington Univ. (St. Louis); MEEBO oligos printed on epoxy slides.
Description
of MEEBO oligos
MEEBO
data sheet
Download Gene File for 2006/2007 Chips produced at
Washington Univ. (St. Louis); MEEBO oligos printed on epoxy slides.
Description of
MEEBO oligos
MEEBO data sheet
Download Gene File for 2005/2006 Chips produced at Washington
Univ. (St. Louis); MEEBO oligos printed on epoxy slides.
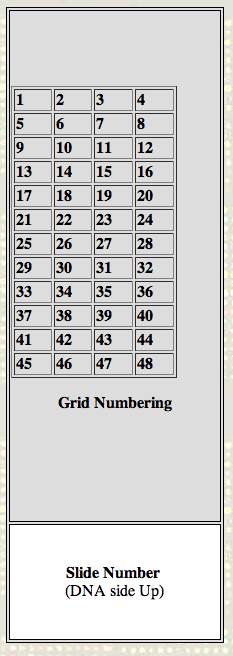
How to orient the spots:
Turn the slide so that it is a tall rectangle with the slide
number etched at the bottom of the slide, facing up. There are 48
grids containing ~38,000 spots.
| 1 |
2 |
3 |
4 |
| 5 |
6 |
7 |
8 |
| 9 |
10 |
11 |
12 |
| 13 |
14 |
15 |
16 |
| 17 |
18 |
19 |
20 |
| 21 |
22 |
23 |
24 |
| 25 |
26 |
27 |
28 |
| 29 |
30 |
31 |
32 |
| 33 |
34 |
35 |
36 |
| 37 |
38 |
39 |
40 |
| 41 |
42 |
43 |
44 |
| 45 |
46 |
47 |
48 |
Grid Numbering
|
Slide Number
(DNA side Up)
|
Within each grid, the 2nd spot is to the right of the 1st
spot; spot #1 is in the top left of each grid.
Basically, once you have the slide oriented, everything
is just like a book...
Download Gene File for 2004/2005
Description of these mouse oligos
Description of Oligo designs in general.
Download Gene File
for 2004-2005 (excel format)
Download Gene File
for 2004-2005 (tab delimited text format)
Download
Gene File for 2004-2005 (gal format)
C. elegans
General Announcement of Long Oligomer-based Spotted Microarrays for the C.
elegans Genome
http://www.genome.wustl.edu/genome/celegans/microarray/ma_gen_info.cgi
Download Gene File for 2007/2008 Chips produced at Washington
Univ. (St. Louis); oligos printed on epoxy slides.
Download Gene File for 2006/2007 Chips produced at Washington
Univ. (St. Louis); oligos printed on epoxy slides.
The Genome Sequencing Center at Washington University has received funding
from NHGRI and HHMI to produce and distribute microarrays for use by C.
elegans investigators. We feel that providing a common resource will enable comparability
of microarray data sets between C. elegans laboratories. The microarrays will
contain long oligomers (nominally 60 mer) that are designed to uniquely represent
each gene in C. elegans (one oligo per gene), placed onto treated glass slides
using split pin technology. Also included on-array will be long oligomers representing
unique E. coli genes, thus allowing investigators to assess relative contamination
of input C. elegans RNA samples with E. coli RNA. We also intend to spot up
to 10 different long oligomers that represent unique Arabidopsis thaliana genes.
RNA samples that correspond to each A. thaliana element can be purchased from
Stratagene (www.Stratagene.com) by end-users, and added to their reverse transcription
reactions during work-up of C. elegans samples. This combination of A.
thaliana oligo elements and RNAs will act as controls for cDNA and labeling reactions,
as well as providing a source of array data normalization that is independent
of sample preparation.
Soybeans (starting fall 2007)
Genelists are emailed directly to end users. Not permitted to post them on
this web site.
The 19kA oligo array contains 19,200 spots of 18,816 unique 70-mer oligos
and 96 oligos spotted 4 times each.
The 19kB oligo array contains 19,200 spots of an additional 18,816 unique 70-mer
oligos and 96 oligos spotted 4 times each.
Annotation
file (19MB).
A standard gal file is provided along with an electronic file providing unique
ID spot locations, 70-mer sequence and the clone ID and Accession number containing
the 70-mer are provided with the slide shipment in Excel format. It is the
responsibility of the users to convert these files to the appropriate format
needed for their own use with their laser scanning equipment.
The oligo arrays have been used in the following publications: Gonzalez and
Vodkin, BMC Genomics 8: 468 (2007).
Tomato (starting fall 2007)
Download Gene File for 2007/2008 and 2008-09 Chips produced
at Cornell Univ.; oligos printed on epoxy slides.
Rice (starting fall 2007)
Gene file
for Fall 2009 (MAGIC Tool ready)
Gene file
for Fall 2009 (Excel file)
Gene file
for Fall 2009 (.gal file)
Spot number one is top left corner.
Spot number two is to
the right of spot number 1 in this orientation.
Zebrafish (starting fall 2007)
If you use 3DNA, you cannot use Array 900 kit, you must use the Array 350
kit. Here are the technical details.
Read Me File (html format)
Zebrafish Gene List (Excel Format)
Zebrafish Gene List (MAGIC Tool Ready Format)
Zebrafish Gene List (.gal Format)
Agilent 2-color protocol (PDF file)
Zebrafish Gene Description File (Excel file, ENABLE MACRO)
GCAT
Home Page
Biology Home Page
© Copyright 2009 Department of Biology, Davidson
College, Davidson, NC 28035
Send comments, questions, and suggestions to: macampbell@davidson.edu