B.S., Davidson College (cum laude)
Watson Fellow, 1984
Ph.D., The Johns Hopkins University
Postdoctoral Fellow, The Johns Hopkins University
Research Associate, Washington University
PEW Visiting Assistant Professor of Biology, Macalester College, MN
Assistant Professor of Biology, Davidson College 1994 - 2000
Visiting Scholar Duke University, Cell and Molecular Biology Program 1999-2000
Associate Professor of Biology, Davidson College 2000 - 2007
Professor of
Biology, Davidson College 2007 to present
Publications:
A. Malcolm
Campbell, Laurie J. Heyer and Christopher J. Paradise. 2014. Integrating
Concepts in Biology. Trunity.com eBook.
The effectiveness of this book was published in 2012 (Barsoum,
et al., CBE Life Sciences Education).
* Book review "Integrating
Concepts in Biology: A Model for More Effective Ways to
Introduce Students to Biology" by K. N. Prestwich and A. M.
Sheehy. 2015. CBE Life Sciences Education.
* MSU validates ICB effectiveness:
Luckie, D.B. et al., 2017. “Integrating
Concepts in Biology Textbook Increases Learning: Assessment
Triangulation Using Concept Inventory, Card Sorting, and MCAT
Instruments, Followed by Longitudinal Tracking.” CBE
Life Sciences Education. Vol 16 (2): article 20.
Ho-Shing, Olivia*, Kin H. Lau*, William Vernon*, Todd T. Eckdahl, and A. Malcolm Campbell. 2012. Assembly of Standardized DNA Parts Using BioBrick Ends in E. coli. Chapter published in “Gene Synthesis: Methods and Protocols”. Metods in Molecular Biology. Volume 852. Jean Peccoud editor. Humana Press. New York, NY. Pages 61 – 76.
A. Malcolm Campbell and Laurie J. Heyer. 2006. Discovering Genomics, Proteomics & Bioinformatics Second Edition. Published jointly by Cold Spring Harbor Laboratory Press and Benjamin Cummings. 447 pages. ISBN 0-8053-8219-4
A. Malcolm. Campbell and Laurie J. Heyer. 2006. Instructor’s Guide to “Discovering Genomics, Proteomics & Bioinformatics”. Benjamin Cummings. in press.
A. Malcolm Campbell and Laurie J. Heyer. 2002. Discovering Genomics, Proteomics & Bioinformatics. Published jointly by Cold Spring Harbor Laboratory Press and Benjamin Cummings. 352 pages. ISBN 0-8053-4722-4
A. Malcolm. Campbell. 2002. Instructor’s Guide to “Discovering Genomics, Proteomics & Bioinformatics”. Benjamin Cummings. 136 pages. ISBN: 0805347267
Abstracts: ( * denotes undergraduates as co-authors)
Workshops and Presentations


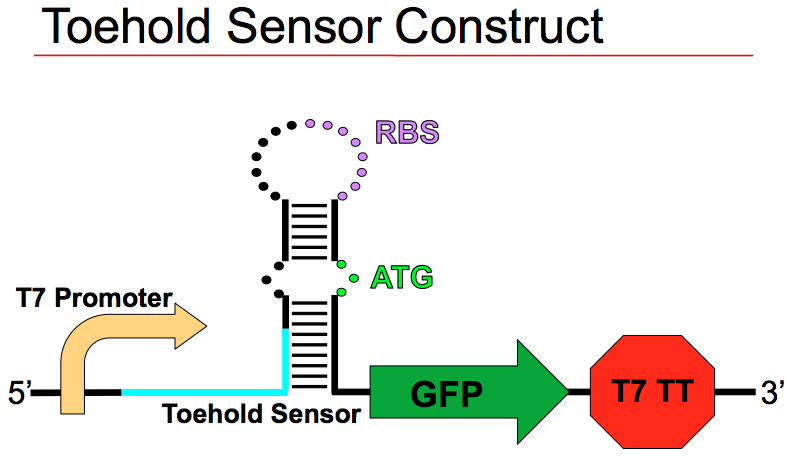


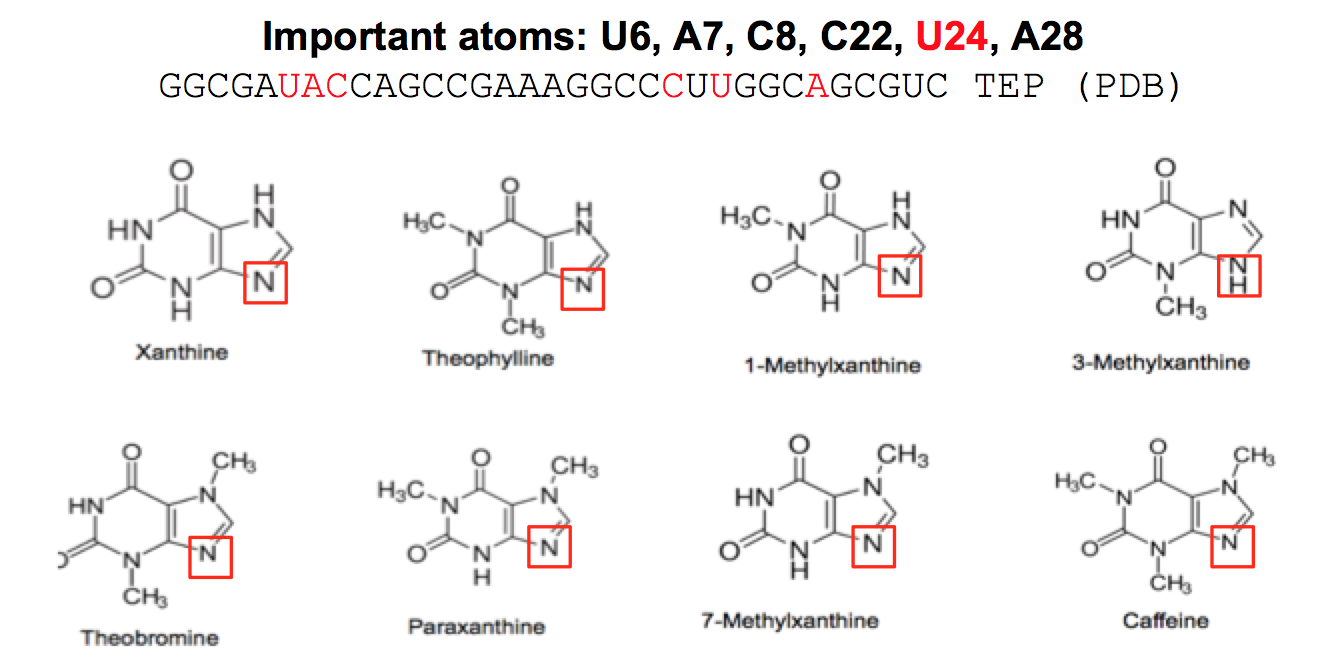
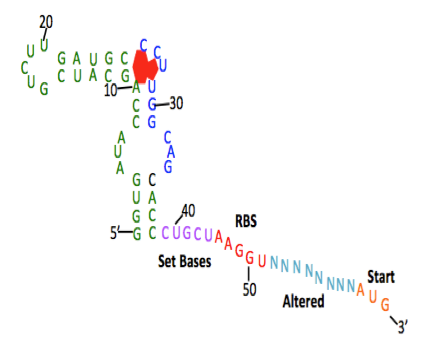
Above: Summer 2015 Davidson College research students. Below: Screen shot from PPT presentation where students discussed the six target bases in the theophylline (TEP) aptamer. These six bases are the key to this RNA aptamer binding to theophylline and they wanted to mutate the bases in order to find a new sequence that could either bind theophylline or one of the other seven members of the xanthine family.
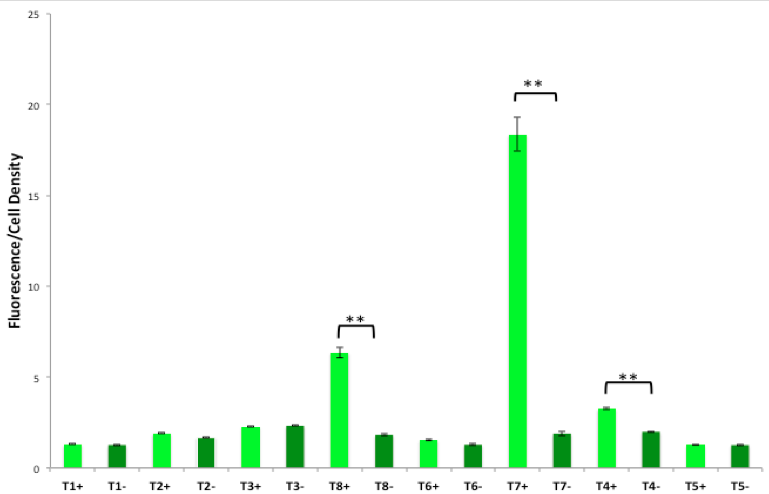
Above: Combined teams of summer students from both campuses. Below: Screen shot of a figure from the 2015 Programmed Evolution paper showing how cells identified genetic constructs that produced the optimal amount of theophylline from caffeine as a substrate.



Summer 2010 students (counter clockwise): Anvi, Jamela, Stephen, Linda (German visiting student), Drs. Campbell and Heyer, Katie, Steph, Tom, Tori (Woodlawn rising junior in high school), Eugene, Nitya, Keila (NC A&T visiting student)


2008 Davidson
College iGEM Team of Math and Biology Students
Every Friday is Hawaiian
shirt day in the town of Davidvson.
Back row: Max, James, Samantha.
Middle row: Kelly, Erin, Dr. Heyer, Kristi, Pallavi.
Front row: Dr. Campbell, Karlesha, Madeline.
Two seniors from the class of 2009 conducted research with me. Samantha Simpson was a genomics major through the Center of Interdisciplinary Studies. Will DeLoache was a biology major who graduated with Honors in Biology.

Samantha and Will, class of 2009
|
Science
Update (AAAS)
(June 25, 2008) |
||||
|
ComputerWorld
(3 June, 2008) |
MSNBC
(2 June, 2008) |
NetworkWorld
(2 June, 2008) |
Scientific
American
(30 May, 2008) |
Digg.com
(29 May, 2008) |
|
Discovery
Channel
(28 May, 2008) |
Skeptic's
Guide to the Universe podcast
(25 May, 2008) |
NanoWerk
(23 May, 2008) |
NPR
Science Friday
(23 May, 2008) |
Informatica
in Brazil
(23 May, 2008) |
|
Buzz
Out Loud podcast
(23 May, 2008) |
Davidson
College News
(22 May, 2008) |
SyntheticBiology.net
(22 May, 2008) |
Delfi
in Estonia
(22 May) |
iTnews
in Australia
(21 May, 2008) |
|
Gizmag
(21 May, 2008) |
Gizmodo
(21 May, 2008) |
Romanian
News
(21 May, 2008) |
Science
Centric
(21 May, 2008) |
OnLineWebLibrary
(21 May, 2008) |
|
Science
Daily
(21 May, 2008) |
Machines
Like Us
(21 May, 2008) |
Sott.Net
(21 May, 2008) |
Russian
Newspaper #1
(21 May, 2008) |
Russian
Newspaper #2
(21 May, 2008) |
|
ThinkGene
blog
(20 May, 2008) |
Genome
Technology Online
(20 May, 2008) |
Top
News India
(20 May, 2008) |
Former
Soviet Union
(20 May, 2008) |
Daily
Science News
(20 May, 2008) |
|
Yahoo
News
(20 May, 2008) |
Medical
News Today
(20 May, 2008) |
Daily
India
(20 May, 2008) |
Khabar
Express
(20 May, 2008) |
innovations
report
(20 May, 2008) |
|
SmasHits
(20 May, 2008) |
AlphaGalileo
(20 May, 2008) |
London
Daily Telegraph
(20 May, 2008) |
Science
News
(19 May, 2008) |
First
Science
(19 May, 2008) |
|
Lab
Spaces.Net
(19 May, 2008) |
Genetic
Engineering News
(19 May, 2008) |
Bio-Medicine
News
(19 May, 2008) |
BioCompare
(19 May, 2008) |
EurekaAlert!
(19 May, 2008) |
Two seniors (2008) conducted their Honors research with me in the field of synthetic biology. Andrew Martens designed a method to bring the E. coli tryptophan anti-terminator into the BioBrick Registry of parts. This part allows transcription to be regulated by the concentration of the amino acid tryptophan.
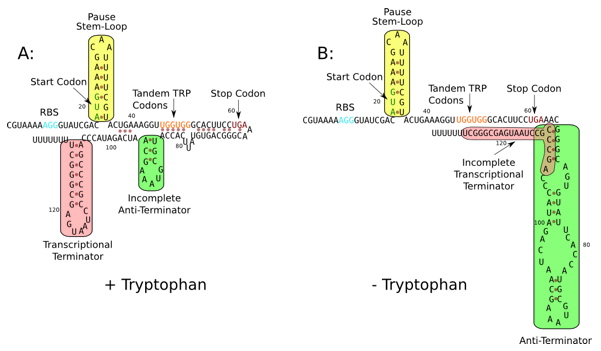
Erin Zwack designed modified anti-switches for E. coli to regulate protein production at the translation step.
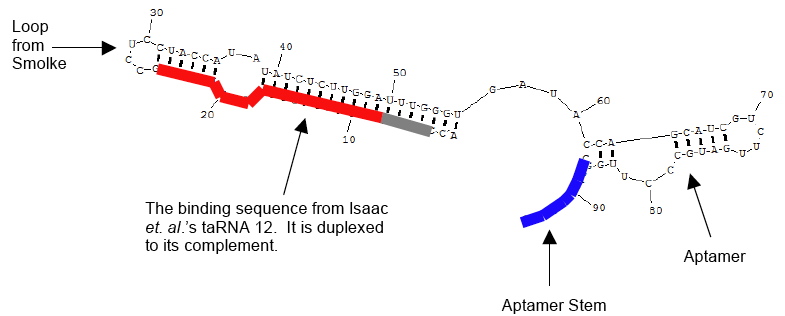
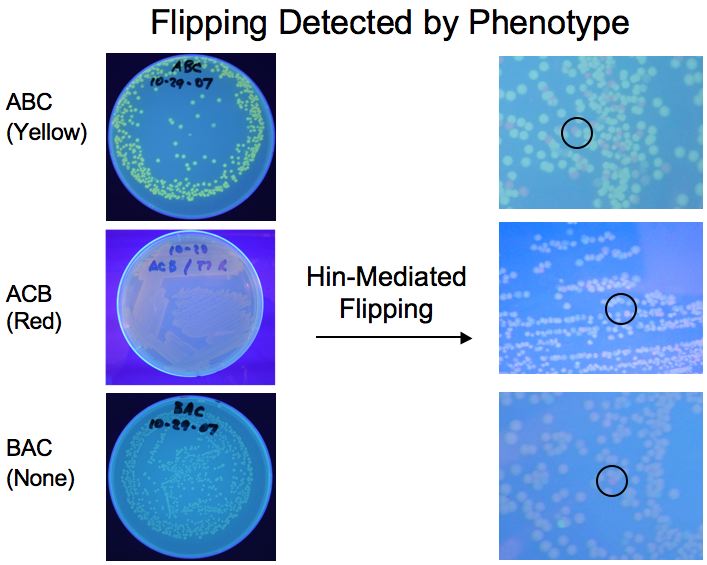
We all learned a lot, but in the end it appears we
were successful in our project. Cells flipped the DNA
and presented the appropriate phenotypes. You can
download the PPT
slide show and the PPT
poster for more details.
In the summer of 2006, 3 students (Erin, Sabriya, and Lance) began an exciting new project to construct an E. coli computer. You can read more about their work from the Davidson team's wiki page or viewing their iGEM 2006 presentation. The key was to integrate biology and mathematics. This project will continue in the fall of 2006 when Samantha and Karmella continue the summer work as we prepare for the 2006 iGEM competition at MIT along with our collaborators at Missouri Western State University. One product of the summer was a new web site that helps investigators synthesize short genes from a series of oligos. The web site, created by Lance, facilitates choosing the right oligos to make. Davidson won prizes in 4 categories at the 2006 iGEM competion. You can read a summary of their successes here. During the sprin semester, Karen Acker inserted Hix sites inside GFP and TetA(C) genes [read her paper] Bruce Henschen produced Hix sites that would allow only one flip [read his paper].
Students in my Genomics Methodology Course (spring 2005 & 2006) have developed what we call a "Teaching Chip" which permits students to design and print their own DNA microarray. Then using very controlled conditions that do not require the isolation of RNA or genomic DNA, they probe their own microarray and detect the signal using MAGIC Tool. This process is very robust and allows students to learn all the critical steps involved in microarray experiments, including data analysis.

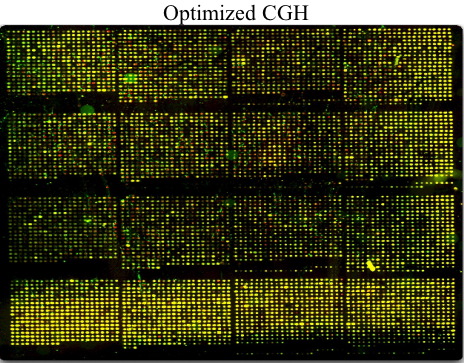
Jackie Ryan '06 developed student friendly protocols for performing comparative genome hybridization experiments. This method uses labeled genomic DNA from two different strains as probes on a yeast DNA microarray. Most spots will be yellow, but aneuploidy (duplicated or deleted DNA) will appear as either red or green spots on a microarray. This work will be continued using evolved strains of yeast induced to undergo aneuploidy by selection under limited glucose.
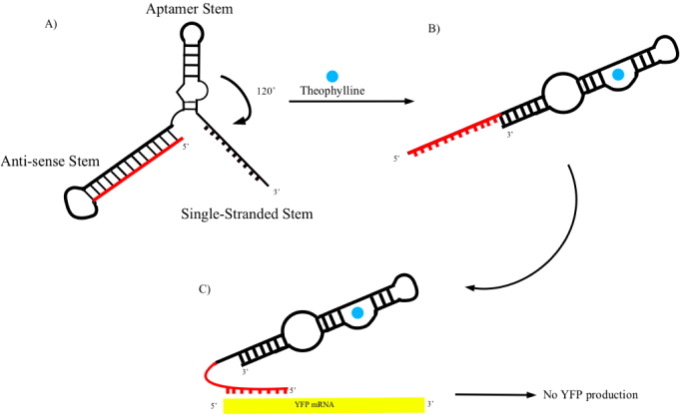
Matt Gemberling '06 has designed and built antiswitches to determine if this new method for regulating eukaryotic translation can be adapted for use in E. coli. Matt's work was part of a larger project in synthetic biology composed of six students (the Synth-Aces) who competed in iGEM2005 at MIT.
Laurie Heyer's students continue to improve the free software for DNA microarray data processing and analysis. This software is called MAGIC Tool (MicroArray Genomic Imaging and Clustering) and works on all computer platforms.
For the 2004-2005 year, Megan McDonald will continue her biophysics major research in collaboration with Dr. Dan Boye of the Davidson Physics Dept. Megan is using optical tweezers to determine the physical topology of DNA printed on glass slides. I am also beginning a project to simplify the DNA microarray protocol enough for high school students to perform exercises that introduce them to the methodology.
In 2003-2004, Gray Lyons completed his honors research project. The lab work was performed at the University of Alabama at Birmingham.Danielle Choi performed two pilot studies with DNA microarrays. In collaboration with Dr. Jef Boeke at Johns Hopkins University, she produced the first yeast bar code DNA microarrays printed on glass slides and worked out hybridization conditions for the probes. She also produced microbial identification DNA microarrays in collaboration with Dr. Brad Goodner at Hiram College. I also helped produce an animation on DNA microarrays.
During 2002-2003, Emily Oldham, John Kogoy and Dan Pierce conducted research in my lab. Emily and Dan worked on improving the signal to noise ratio of DNA microarrays. John conducted some preliminary studies in proteomics.
In the 2001-2002 academic year, Emily Oldham began her genomics concentration through Davidson's Center for Interdisciplinary Studies under the guidance of Laurie Heyer and me. Laurie and I also finished the genomics textbook writing project.
I spent
my sabbatical at the University of Washington in Seattle
and the Institute for Systems Biology to genomics and
develop a course for undergraduates, during the 2000-2001
academic year, . This new
course in genomics for undergraduates was offered
for the first time during the fall of 2001. Furthermore, I
have created a new non-profit educational consortium
called the Genome
Consortium for Active Teaching (GCAT).
GCAT
is dedicated to bringing genomic methods into the
undergraduate laboratory curriculum at a reasonable cost.
During
the 1999-2000 academic year, one of my honors
students will be conducting research in the new and
very exciting field of bioinformatics. Rahul Karnik '00 is
a Biology major and is also working on a concentration in
computer sciences. For the summer of 1999, he worked at TIGR
(The Institute for Genomic Research) in Maryland. He is
currently working at a pharmaceutical company as the link
between the biologists and the computer scientists.
I have been conducting research to improve teaching methods. I work with students and use the web to improve the quality of my lecture and lab curricula. The web provides a medium for presenting visual information that cannot be presented any other way.
One student conducted her honors research to develop a project-based semester long laboratory course for Molecular Biology.
I have also redesigned my Molecular Biology curriculum, and as well as Introductory Biology.
I
have developed interactive animations and image maps for
teaching immunology.
For this, I have created "Molecular
Movies"
and "Hyperlinked
Human
Histology".
For
several years, we have been examining proteins involved in
mating of the unicellular green alga called Chlamydomonas.
We have generated two mutant strains (iso1
and pmh1 )
that display abnormal mating phenotypes. Several Davidson
College students have helped identify the molecular cause
for the pmh1 mutation and this work is being
submitted for publication (see virtual
posters below).
Another
area of interest in my lab has been the purification and
characterization of NADP+-dependent enzymes
from Chlamydomonas (click
here for a list of examples).This work has been
conducted in collaboration with Dr. John Williamson, also
in the Biology Dept. We have had several student conduct
research in this area.
During the 1998-99 academic year, one of my students wanted to isolate the protein(s) that bind to a new drug being tested for use in treating Alzheimer's disease. His honors thesis research involved Dr. Brown of the Chemistry Dept., as well as Dr. Williamson and me. This new area of investigation was awarded first prize at the Collegiate Academy of the North Carolina Academy of Sciences 1999 annual meeting.
Links to Virtual Posters:
Physical Properties of DNA on Microarrays (work in progress)
Examining Accuracy and Precision of DNA Microarrays (1 MB PowerPoint file)
Exploring the Use of Plasmids in DNA Microarray Technology (1 MB PowerPoint file)
Proteomics Modules (web site - requires chime plugin)
Helping Students Discover Genomics, Proteomics and Bioinformatics. (4.4 MB PDF file)
Comparison
of the Use of Plasmid and PCR DNA on Microarray
Chips (1 MB
PDF file)
Current Teaching Interests:
| Bio113:
Introductory Biology (Information, Evolution, Cells) |
Bio114
: Introductory Biology (Cells, Emergent Properties, Homeostasis) |
| Bio111:
Introductory Biology (Cell and Molecular) |
Bio111
Lab: Introductory Biology (Cell and Molecular) |
| Bio304: Molecular Biology | Bio304 Lab: Molecular Biology Lab |
| Bio309: Genomics, Proteomics and Systems Biology | Bio343 Laboratory Methods in Genomics |
| Bio307: Immunology (spring 2006 my last time) | Concentration
in Genomics (established fall 2004) |
| Seminar
in Synthetic Biology |
Group Investigation in Synthetic Biology |
Send comments, questions, and suggestions to: macampbell@davidson.edu
or (704) 894 - 2692 ©
Copyright 2013 Department of Biology, PO Box 7118,
Davidson College,
Davidson, NC 28035